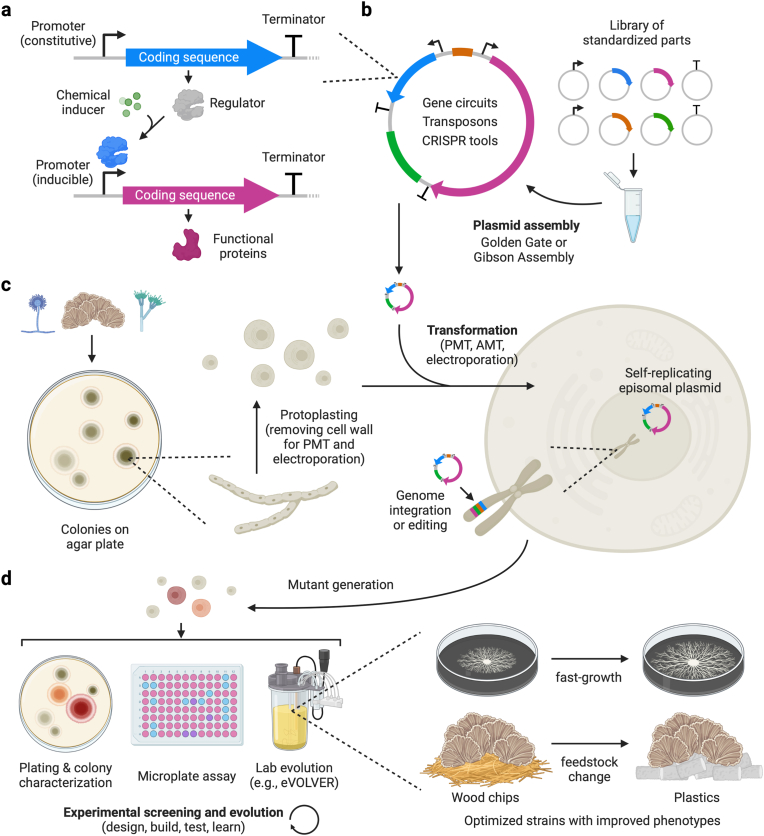
Fig. 3.
The synthetic biology workflow for strain engineering and optimization. (a) A transcription unit has three components: a promoter, the coding sequence, and a terminator. An inducible promoter can be activated or inactivated by a regulator protein that undergoes conformational changes upon binding with small molecules. (b) Part plasmids hosting promoters, coding sequences, terminators, selection markers, and origin of replication are assembled into a single plasmid using modular cloning strategies. (c) Transformation methods such as PEG-mediated transformation (PMT), Agrobacterium-mediated transformation (AMT), and electroporation transfer the assembled plasmid or linear payload into the fungal cells. The introduced DNA can either self-replicate independently or homologously recombine with the host chromosome. Genetic payloads on an episomal plasmid or integrated into the host genome drive the expression of exogenous proteins or modify the genome, generating mutants with various phenotypes. (d) Researchers often use colony morphology characterization, microplate assay, and continuous evolution techniques to benchmark the mutants. Depending on the selection or evolution methods, the optimized strains could have desirable traits such as fast growth, feedstock change, or higher protein secretion yield.