Figure 1 |. Geographical locations and genetic variants across populations.
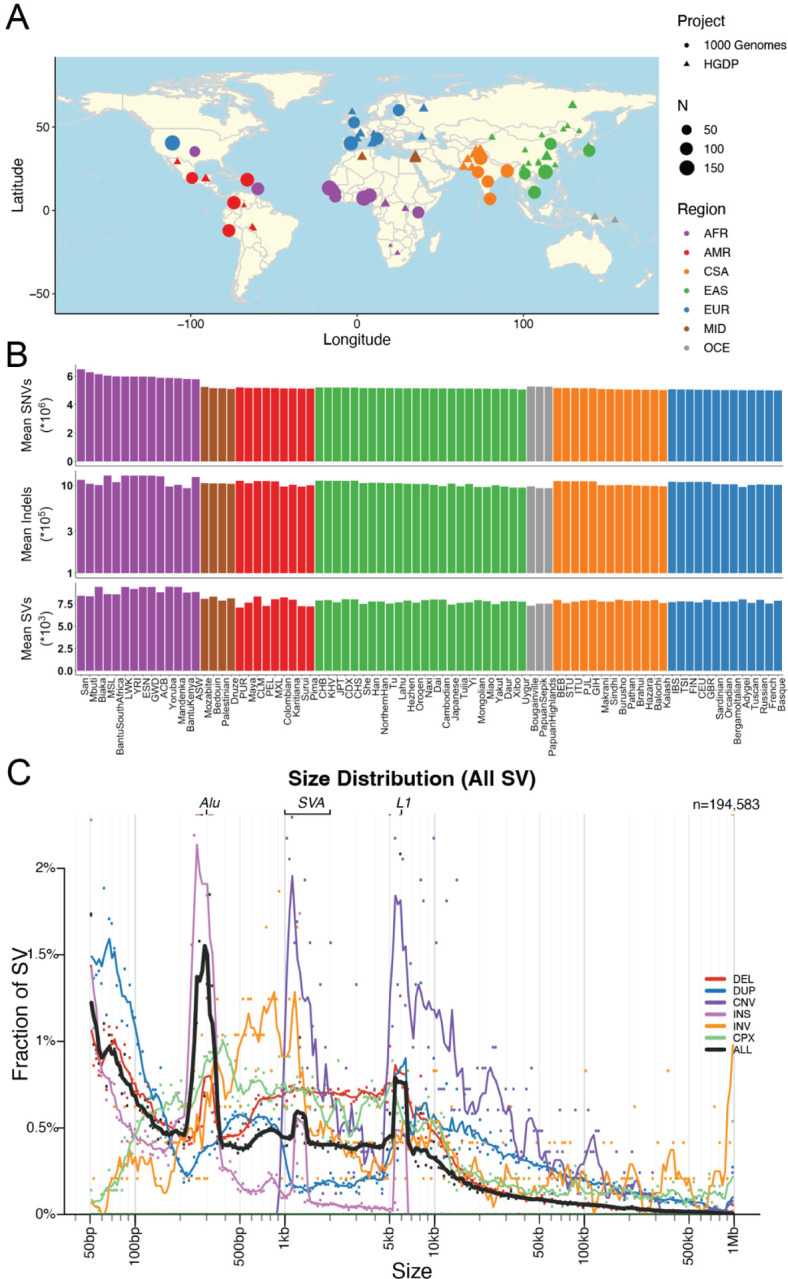
A) Global map indicating approximate geographical locations where samples were collected. Coordinates were included for each population originating from the Geography of Genetic Variants browser as well as meta-data from the HGDP (Bergström et al. 2020; Marcus and Novembre 2017). B) Mean number of SNVs (top), indels (middle), and SVs (bottom) per individual within each population. Colors are consistent with geographical/genetic regions in A-B), as follows: AFR=African, AMR=admixed American, CSA=Central/South Asian, EAS=East Asian, EUR=European, MID=Middle Eastern, OCE=Oceanian. C) Sizes of SVs decay in frequency with increasing size overall with notable exceptions of mobile elements, including Alu, SVA, and LINE1. Abbreviations are deletion (DEL), duplication (DUP), copy number variant (CNV), insertion (INS), inversion (INV), or complex rearrangement (CPX).
