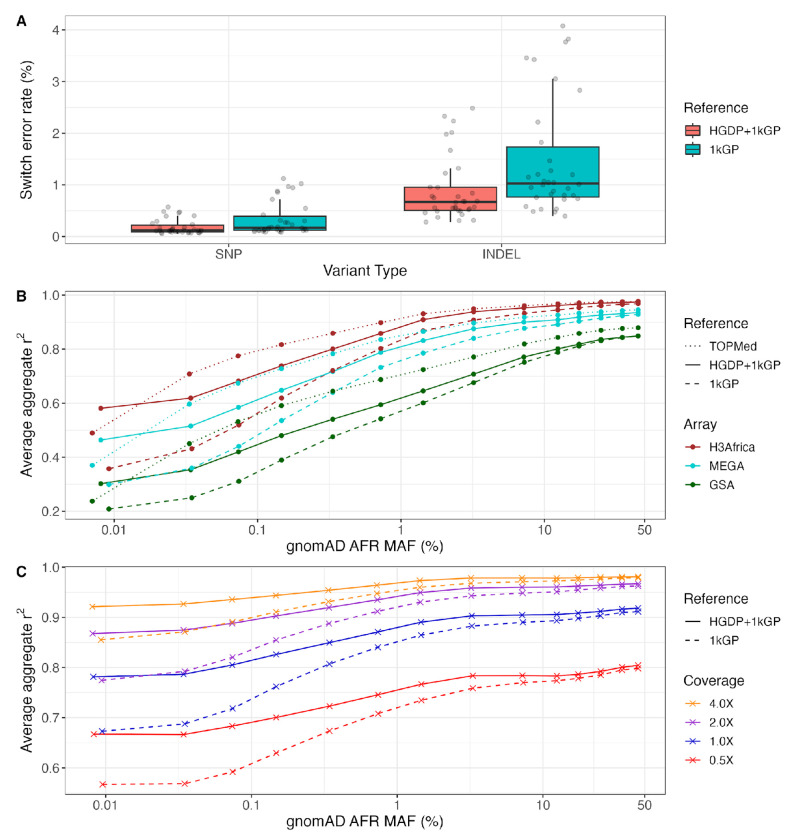
Figure 5 |. Phasing and imputation accuracy are improved across data generation strategies compared to existing reference panels.
A) Switch error rates for SNPs and indels in a truth set of 34 HGSVC2 genomes when using HGDP+1kGP versus 1kGP reference panels for phasing. B-C) Imputation performance as a function of minor allele frequency (MAF) for AFR in gnomAD v3.1 data using TOPMed, HGDP+1kGP, and 1kGP reference panels in B) SNP array and C) low-coverage sequencing data. Aggregate r2, which is the correlation between the imputed dosages and high coverage “truth” genotype calls, was computed in MAF bins and averaged across chromosomes 1-22. The validation set is composed of 93 AFR individuals sequenced at 30X coverage (Martin et al. 2021).