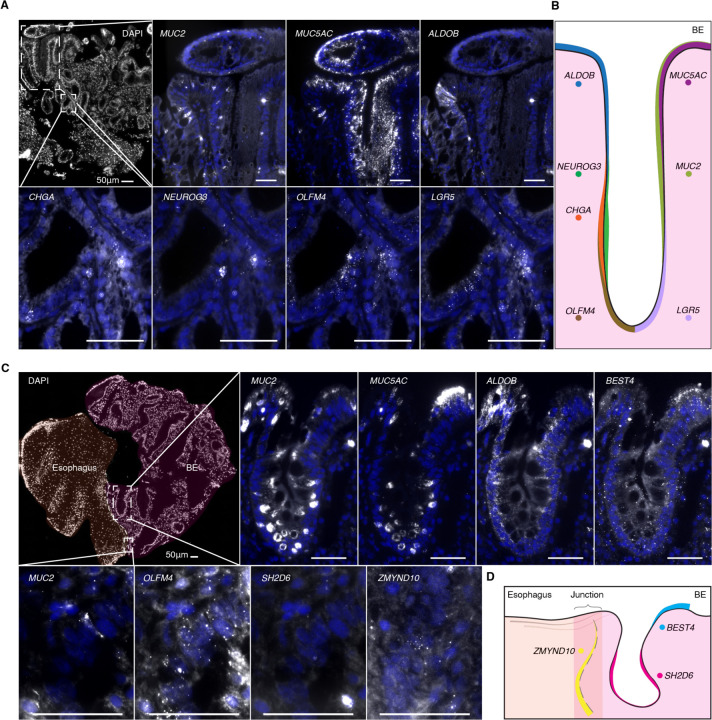
Fig. 3. Spatial location of Barrett’s esophagus cell types.
(A) Multiplexed HCR RNA FISH of fresh frozen Barrett’s esophagus sections for MUC2 (goblet), MUC5AC (foveolar), ALDOB (enterocyte), CHGA (enteroendocrine), NEUROG3 (enteroendocrine progenitor), OLFM4 (stem), and LGR5 (stem) with DAPI counterstain. Scale bars, 50 μm. DAPI staining is displayed in blue. All genes were detected in sections from at least two patients. (B) Schematic depicting the general location of key marker genes within Barrett’s esophagus glands. (C) Multiplexed HCR RNA FISH of fresh frozen tissue sections from the junction of Barrett’s esophagus and squamous esophagus for markers of newly discovered rare cell types, as well as typical marker genes from (A): SH2D6 (tuft), ZMYND10 (ciliated), and BEST4. Scale bars, 50 μm. DAPI staining is displayed in blue. All genes were detected in sections from at least two patients, with the exception of ZMYND10. (D) Schematic depicting the general location of rare-cell markers within Barrett’s esophagus tissue.