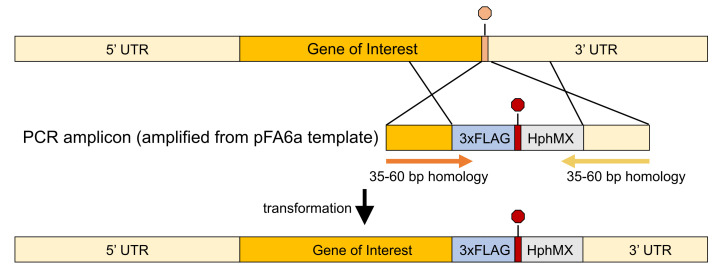
Figure 2. Overview of primer design for yeast transformation.
Primers are designed with the forward strand being homologous to the 3’ end of the gene of interest (GOI) just before the endogenous stop codon and the reverse strand being homologous with a segment immediately following the stop codon. The primers anneal to a region of a pFA6a plasmid (e.g., pTF262; see Materials) that encode a 3xFLAG tag and selectable marker (in this case, hygromycin). Following PCR amplification and yeast transformation, the cells now express a 3xFLAG-tagged version of the GOI and a selectable marker.