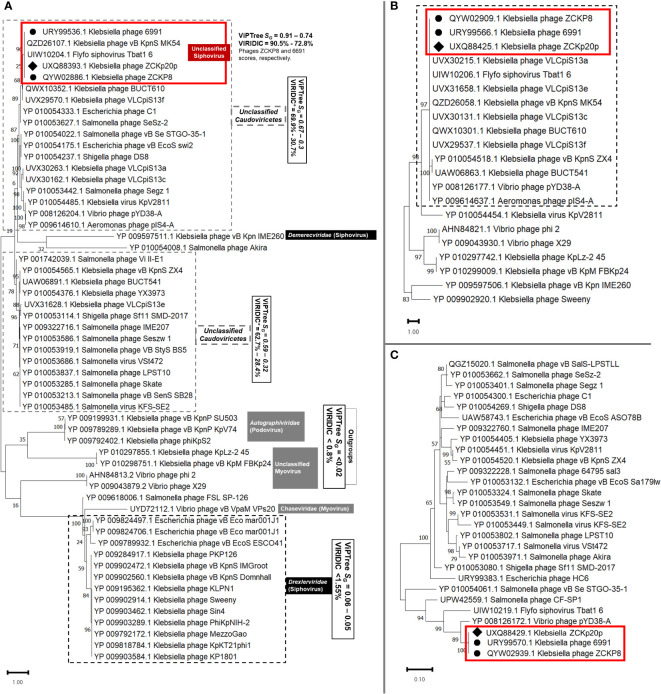
Figure 12.
Viral conserved signature protein-based phylogenetic tree. (A) A phylogenetic tree for the evolutionary relatedness of the terminase large subunits of phage vB_Kpn_ZCKp20p to its corresponding proteins in other phages. (B) Phylogenetic tree of the major capsid protein of vB_Kpn_ZCKp20p and its orthologs in closely related phages. (C) Phylogenetic tree of the DNA polymerase III beta subunit of phage vB_Kpn_ZCKp20p and its orthologs in other phages. Phages included in the evolutionary analysis were chosen based on the closest relatives according to the proteomic tree and intergenomic similarity analysis. The phylogenetic distance was inferred by the Maximum likelihood method and supported with a bootstrap of 1000 replicates, and the trees were constructed by MEGA11. Phage vB_Kpn_ZCKp20p clusters were outlined in red based on the protein homology in both trees of the highest log likelihood (-12536.16, -5522.81 and -1839.71, respectively).