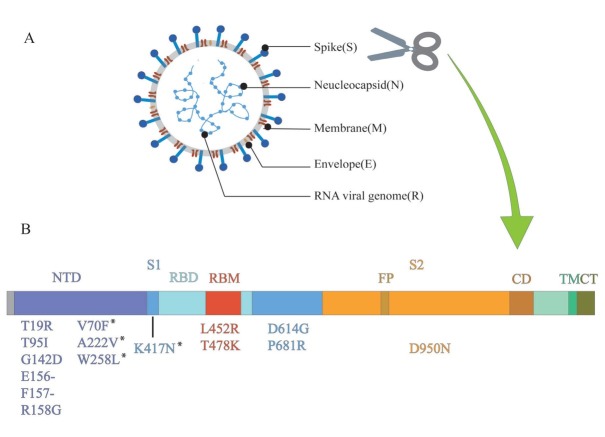
Figure 1.
Structure of SARS-CoV-2 and mutations of delta variant. (A) SARS-CoV-2 forms coated spherical particles with diameters of 100–160 nm. It contains a 27–32 kb positive ssRNA genome. Envelope glycoproteins spike protein (S), envelope (E), membrane (M), and nucleocapsid (N) are encoded by 3' terminal genome. (B) Spike protein is critical for the virus to invade the host, which is also a key focus in the research on SARS-CoV-2. S1 and S2 subunits and a transmembrane domain constitute a spike protein. Scissor cuts at the boundary of S1 and S2. RBD containing the core-RBD and RBM, and NTD are on S1, which undertakes the role of binding to the host cell receptor, especially RBD, while S2 consists of FP, CD, and CT, contributing to membrane fusion. Mutations of delta variant are marked in the figure. ssRNA: single-stranded RNA; NTD: amino-terminal domain; RBD: receptor-binding domain; RBM: receptor-binding motif; FP: fusion peptide; CD: connecting domain; TM: transmembrane domain; CT: cytoplasmic tail; SARS-CoV-2: severe acute respiratory syndrome novel coronavirus. *Detected in some sequences of delta variant, but not all.