Figure 2. Differentially expressed genes (DEGs) and their functional categories reveal transcriptomic signatures of inflammatory aortic aneurysms.
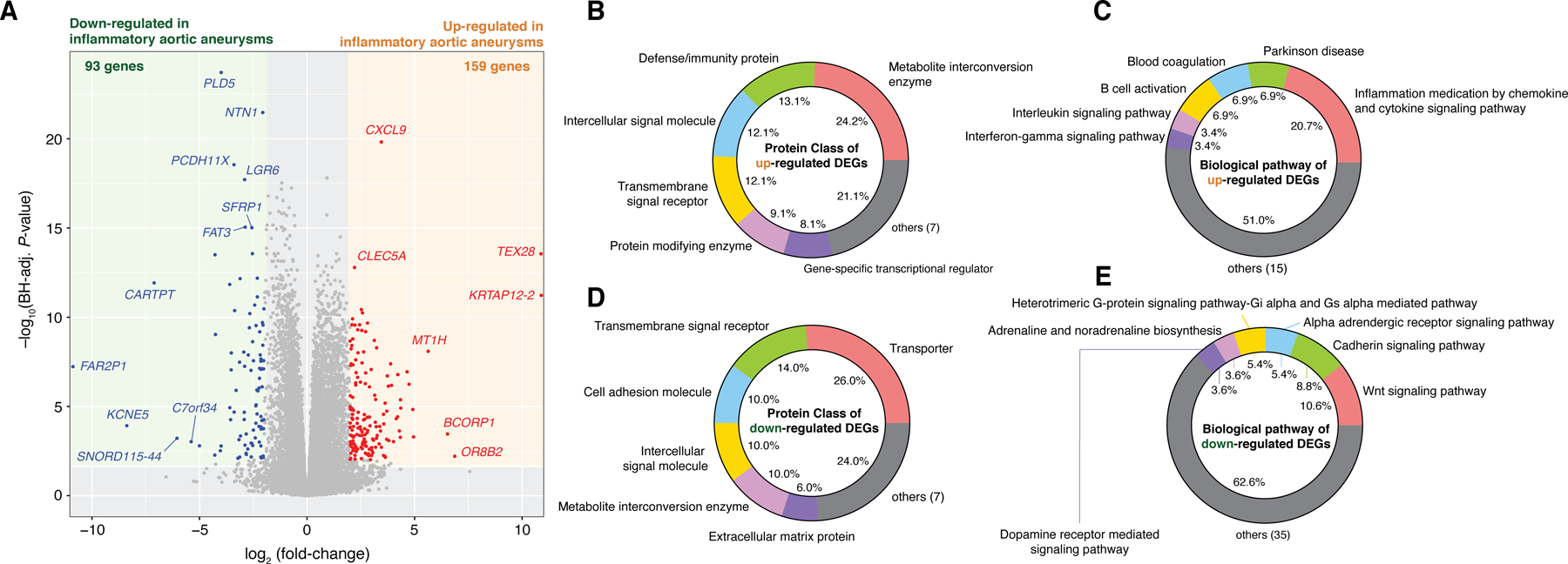
(A) A total of 159 and 93 genes were identified to be significantly upregulated and downregulated, respectively, in inflammatory aortic aneurysms (Benjamini-Hochberg adjusted P-value < 0.01 and |log2(fold-change in mean expression values)| > 2). (B) Protein classes and (C) biological pathways of the upregulated DEGs. (D) Protein classes and (E) biological pathways of the downregulated DEGs. DEGs and fold-changes were calculated by DEseq2 (v1.30.0) while controlling for ACE/ARB use, statin use, history of other RD. Functional classification of protein class and biological pathways was performed using the PANTHER Database (v16.0).
