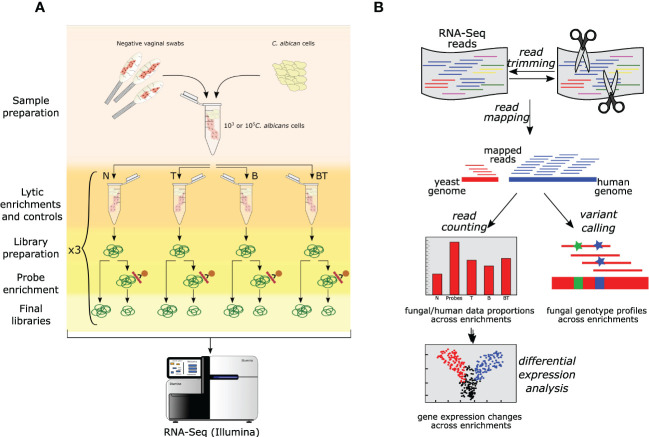
Figure 1.
Overall experimental design and data analysis pipeline used in this study. (A) Schematic representation of the experimental setup. A pool of Candida-negative vaginal samples was spiked with different loads of C. albicans cells. Spiked samples further underwent different enrichments and sequencing. Different lytic enrichment approaches are: N - No thiolutin and no lytic enrichment; T - Thiolutin and no lytic enrichment; B - No thiolutin and buffer RLT + β-mercaptoethanol lytic enrichment; BT - Thiolutin and Buffer RLT + β-mercaptoethanol lytic enrichment. (B) Schematic RNA-Seq data analysis workflow employed in this study (see Materials and Methods for details).