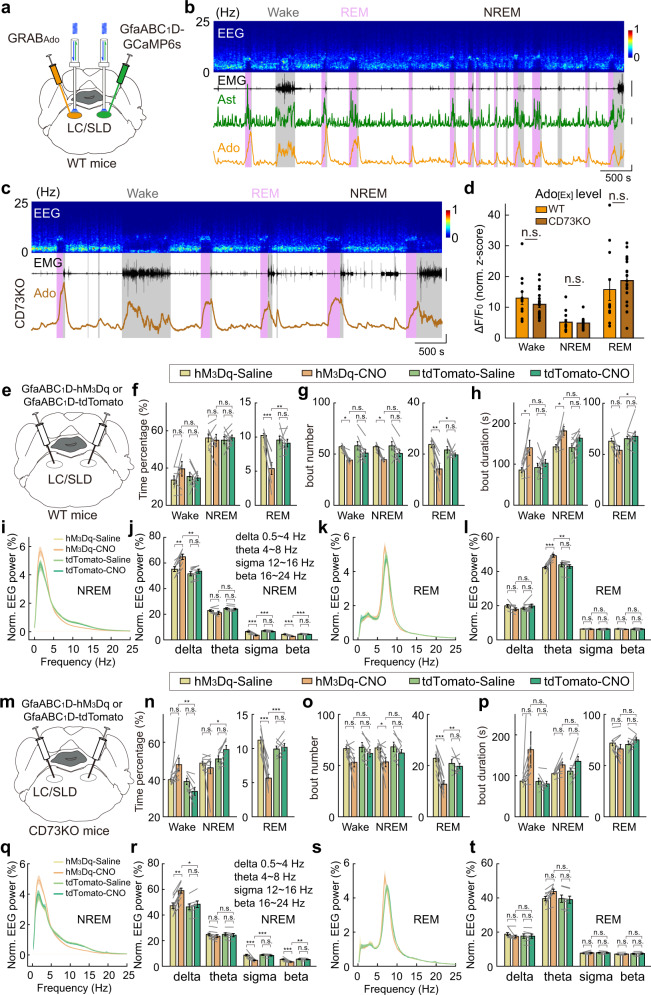
Fig. 8. Sleep–wake regulation by astrocytes in the brainstem.
a Schematic diagram depicting fiber photometry recording of extracellular adenosine level and population Ca2+ signal of astrocytes in LC/SLD region of the brainstem during the sleep–wake cycle. b Top to bottom, EEG power spectrogram, EMG (scale, 1 mV), GCaMP fluorescence (scale, 1 z-score), and GRABAdo fluorescence (scale, 1 z-score). c Fiber photometry recording of extracellular adenosine level in LC/SLD in CD73-KO mice during the sleep–wake cycle. d GRABAdo signal in WT and CD73-KO mice in different brain states. Each line represents data from one recording. n = 12 sessions from 5 mice in the WT group; n = 20 sessions from 6 mice in the CD73-KO group. Wake: P = 0.52; NREM: P = 0.47; REM: P = 0.22; Wilcoxon rank-sum test. e–l Same as Fig. 3g–n, respectively, except that experiments were performed in LC/SLD. hM3Dq group, n = 7 mice; tdTomato group, n = 8 mice. The statistical method used was two-way repeated measures ANOVA, followed by Tukey’s post hoc multiple comparison test. In f, hM3Dq-CNO vs tdTomato-CNO: Wake, P = 0.47; NREM, P = 0.97; REM, **P = 0.0024. hM3Dq-Saline vs hM3Dq-CNO: Wake, P = 0.34; NREM, P = 0.98; REM, ***P < 0.001. tdTomato-Saline vs tdTomato-CNO: Wake, P = 0.99; NREM, P = 0.97; REM, P = 0.94. In g, hM3Dq-CNO vs tdTomato-CNO: Wake, P = 0.35; NREM, P = 0.42; REM, *P = 0.045. hM3Dq-Saline vs hM3Dq-CNO: Wake, *P = 0.030; NREM, *P = 0.034; REM, **P = 0.0011. tdTomato-Saline vs tdTomato-CNO: Wake, P = 0.27; NREM, P = 0.25; REM, P = 0.70. In h, hM3Dq-CNO vs tdTomato-CNO: Wake, P = 0.12; NREM, P = 0.41; REM, *P = 0.045. hM3Dq-Saline vs hM3Dq-CNO: Wake, *P = 0.021; NREM, *P = 0.021; REM, P = 0.28. tdTomato-Saline vs tdTomato-CNO: Wake, P = 0.88; NREM, P = 0.19; REM, P = 0.95. In j, hM3Dq-CNO vs tdTomato-CNO: delta, **P = 0.0021; theta, P = 0.088; sigma, ***P < 0.001; beta, ***P < 0.001. hM3Dq-Saline vs hM3Dq-CNO: delta, **P = 0.0085; theta, P = 0.46; sigma, ***P < 0.001; beta, ***P < 0.001. tdTomato-Saline vs tdTomato-CNO: delta, P = 0.81; theta, P = 0.97; sigma, P = 0.53; beta, P = 0.58. In l, hM3Dq-CNO vs tdTomato-CNO: delta, P = 0.38; theta, **P = 0.0013; sigma, P = 0.98; beta, P = 0.89. hM3Dq-Saline vs hM3Dq-CNO: delta, P = 0.36; theta, ***P < 0.001; sigma, P = 0.97; beta, P = 0.73. tdTomato-Saline vs tdTomato-CNO: delta, P = 0.67; theta, P = 0.84; sigma, P = 0.93; beta, P = 1.0. m–t Same as e–l, respectively, except that experiments were performed in CD73-KO mice. hM3Dq group, n = 9 mice; tdTomato group, n = 7 mice. The statistical method used was two-way repeated measures ANOVA, followed by Tukey’s post hoc multiple comparison test. In n, hM3Dq-CNO vs tdTomato-CNO: Wake, **P = 0.0020; NREM, *P = 0.023; REM, ***P < 0.001. hM3Dq-Saline vs hM3Dq-CNO: Wake, P = 0.064; NREM, P = 0.80; REM, ***P < 0.001. tdTomato-Saline vs tdTomato-CNO: Wake, P = 0.40; NREM, P = 0.40; REM, P = 0.98. In o, hM3Dq-CNO vs tdTomato-CNO: Wake, P = 0.32; NREM, P = 0.34; REM, **P = 0.0077. hM3Dq-Saline vs hM3Dq-CNO: Wake, P = 0.051; NREM, *P = 0.050; REM, ***P < 0.001. tdTomato-Saline vs tdTomato-CNO: Wake, P = 0.68; NREM, P = 0.67; REM, P = 0.93. In p, hM3Dq-CNO vs tdTomato-CNO: Wake, P = 0.098; NREM, P = 0.80; REM, P = 0.23. hM3Dq-Saline vs hM3Dq-CNO: Wake, P = 0.11; NREM, P = 0.12; REM, P = 0.53. tdTomato-Saline vs tdTomato-CNO: Wake, P = 1.0; NREM, P = 0.13; REM, P = 0.77. In r, hM3Dq-CNO vs tdTomato-CNO: delta, *P = 0.015; theta, P = 0.95; sigma, ***P < 0.001; beta, **P = 0.0015. hM3Dq-Saline vs hM3Dq-CNO: delta, **P = 0.0042; theta, P = 0.81; sigma, ***P < 0.001; beta, ***P < 0.001. tdTomato-Saline vs tdTomato-CNO: delta, P = 0.91; theta, P = 0.98; sigma, P = 0.90; beta, P = 0.96. In t, hM3Dq-CNO vs tdTomato-CNO: delta, P = 1.0; theta, P = 0.18; sigma, P = 1.0; beta, P = 0.85 hM3Dq-Saline vs hM3Dq-CNO: delta, P = 0.72; theta, P = 0.22; sigma, P = 0.95; beta, P = 1.0. tdTomato-Saline vs tdTomato-CNO: delta, P = 1.0; theta, P = 0.99; sigma, P = 0.99; beta, P = 0.99.