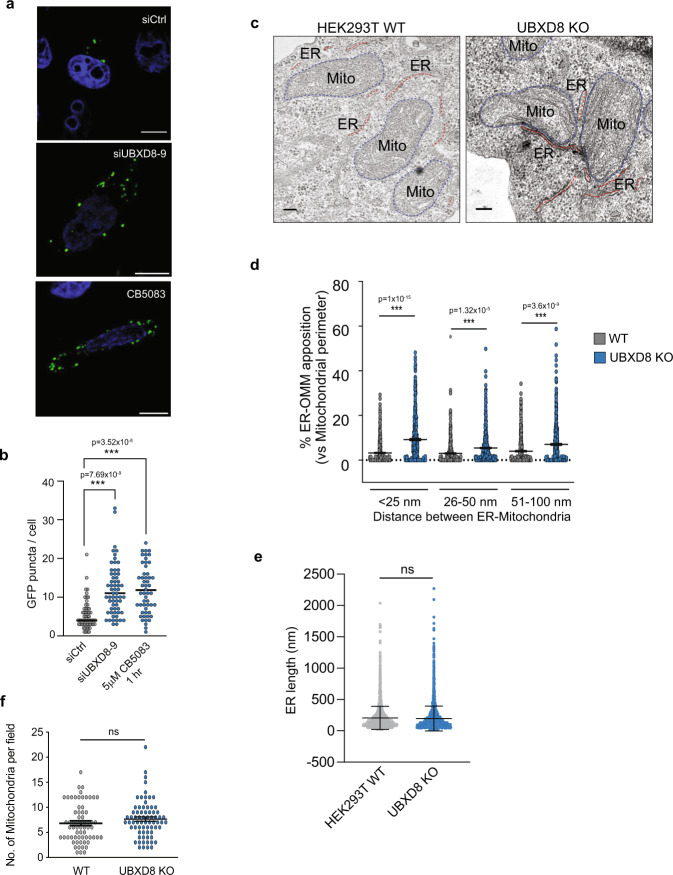
Fig. 3. UBXD8 knock out or p97 inhibition results in increased ERMCS.
a Representative microscopy images showing ERMCS as depicted by formation of fluorescent GFP puncta at ERMCS by split-GFP-based contact site sensor (SPLICS) system comprising OMM-GFP1-11 and ER-short-GFPβ11)27. Scale bar, 10 μm. b Quantification of GFP puncta per cell as a proxy for ERMCS upon siRNA-mediated repression of UBXD8 or p97 inhibition by small molecule inhibitor, CB5083 (5 μM, 1 h). (Cell numbers used for quantification: siControl = 57 cells; siUBXD8#9 = 61 cells; CB5083 = 52 cells, across n = 3 biologically independent samples). c Representative TEM micrographs of wildtype and UBXD8 KO HEK293T cells illustrating contacts between ER (red dotted line) and mitochondria (blue-dotted line). d Quantification of contact length between ER and mitochondria in each genotype from c. e Quantification of ER lengths per field from TEM of wildtype and UBXD8 KO cells from c. f Quantification of number of mitochondria per field from TEM of wildtype and UBXD8 KO cells from c. Measurements in d–f are from n = 3 biological replicates with WT = 50 cells from 65 fields and UBXD8 KO = 53 cells from 71 fields. OMM: Outer mitochondrial membrane. Data are means ± SEM. *, **, ***P < 0.05, 0.01, 0.0001 respectively, one-way ANOVA with Tukey’s multiple comparison test (b, d); two-tailed unpaired t test with Welch’s correction (e, f). Scale bar, 100 nm (c). Source data are provided as a Source data file.