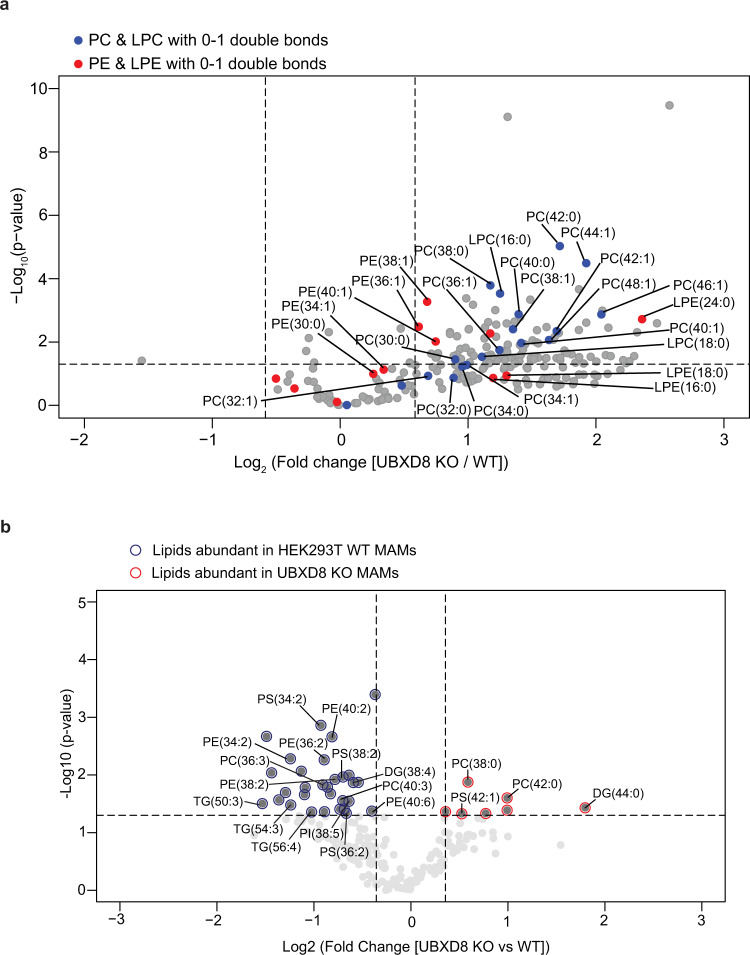
Fig. 5. Quantitative lipidomics of MAMs identifies a role for UBXD8 in regulating fatty acid composition at ERMCS.
a, b Volcano plot of the (−log10 transformed P value vs the log2-transformed ratio of UBXD8 KO: wildtype) total phospho- and neutral lipid species identified using lipidomics of whole cell extracts (a) or MAM fractions (b) of HEK293T cells. PC, LPC species (blue filled circles) and PE, LPE species (red filled circles) with saturated or monounsaturated fatty acid tails are labeled in a. Lipid species with saturated or monounsaturated fatty acid tails (red outline) or polyunsaturated fatty acid tails (blue outline) are labeled in the lipidomics of MAM fractions in b. Lipids were measured by LC-MS/MS following normalization by total protein amount. (n ≥ 3 biologically independent experiments were performed, each with duplicate samples). Statistical analysis was performed on the log transformed relative fold change values (UBXD8 KO relative to WT) using independent two-tailed t tests and Benjamini–Hochberg correction in R stats package (P values are listed in Supplemental Dataset 2). PC phosphatidyl choline, LPC lysophosphatidyl choline, LPE lysophosphatidyl ethanolamine, PE phosphatidyl ethanolamine, PI phosphatidylinositol, PS phosphatidylserine, DG diacylglycerol, TG triacylglycerol.