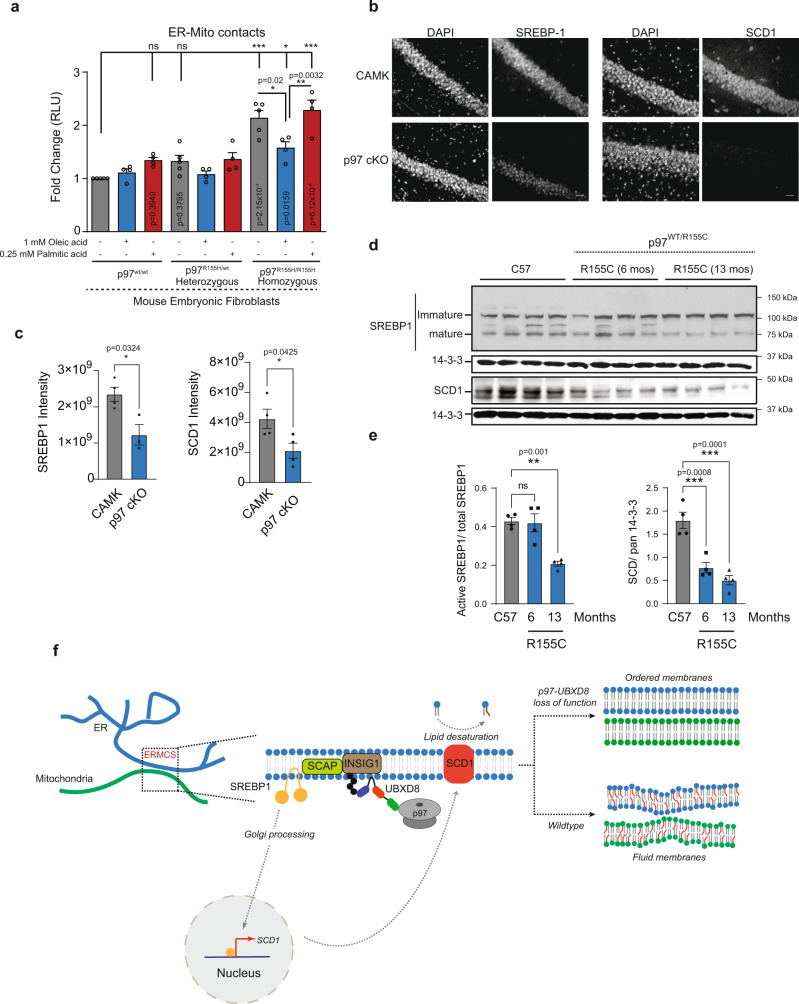
Fig. 8. p97 R155H disease mutation and conditional p97 knock out in mice recapitulates the p97-UBXD8 loss of function effect on ERMCS.
a Split luciferase assay to measure contacts in mouse embryonic fibroblasts with heterozygous or homozygous p97 R155H mutation. Cells were supplemented with indicated concentrations of oleic acid and palmitic acid for 4 h. (For a: n = 5, 4, 4, 5, 4, 4, 5, 4, and 4 biologically independent samples from left to right, respectively). b Representative SREBP1 and SCD1 staining from CA1 regions of 1 month-old control (CAMK2α) and p97 cKO mice (scale bar is 25 μm). c Quantification of fluorescence intensity of images in b. Individual points represent mean ROI intensity from each mouse, 3 or 4 animals per group. d Representative immunoblot for SREBP1 and SCD1 from cortical brain lysates of 12-month-old control (C57), or 6- and 12-month-old p97R155C/WT mice (n = 4 for each group, except for p97 cKO in SREBP1 panel (n = 3)). Pan 14-3-3 was used as housekeeping control. e Quantification of d. The ratio of mature SREBP1 to total SREBP1 is shown. SCD1 intensities are normalized to 14-3-3 levels in each lane. Individual points represent each mouse, 4 animals per group. Data are means ± SEM (*, **, ***P < 0.05, 0.01, 0.0001, respectively. Significance was analyzed by one-way ANOVA with Tukey’s multiple comparison test (a) or Dunnett’s multiple comparison (e) or two-tailed Student’s t test (c). f Model: Schematic of SREBP pathway activation. At ERMCS, UBXD8 recruits p97 to mediate the extraction and degradation of INSIG1 when it is ubiquitylated by the E3 ligase GP78. Loss of INSIG1, which is a negative regulator of SREBP1, allows translocation of cleaved SREBP1 transcription factor into the nucleus to activate expression of lipid desaturases, notably SCD1. Loss of p97-UBXD8 leads to decrease in SCD1 levels and loss of lipid desaturation resulting in membrane order and increased contacts. Source data are provided as a Source data file.