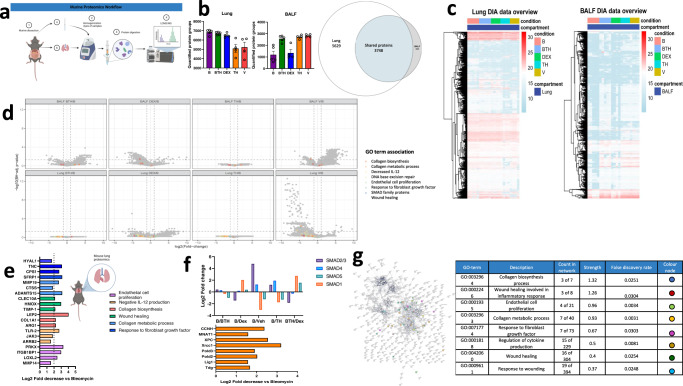
Fig. 6. Proteome of lungs and BALF.
a Experimental workflow showing analysis of protein extracts by mass spectrometry. b Unique and overlapping protein identifications subdivided by treatment condition in lungs and BALF by biological fluid. c Response profile heatmaps displaying protein expression across treatment conditions in each biological fluid. d Volcano plot depicting differentially expressed proteins in BALF (top panel) and lung (bottom panel) with proteins associated with significant gene ontology (GO) terms highlighted in each plot. e, f Specific lung proteins downregulated following Bleo/TH5487 treatment compared to the Bleo alone condition. Proteins highlighted in these plots are specifically associated with e fibrotic-related changes or f SMAD (top panel) and nucleotide base repair (bottom panel). g Network analysis (StringDB) comparing the Bleo/Bleo+TH treatment condition displaying significant biological GO terms, protein counts in the network, strength, and FDR. For LCMS/MS analyses, murine samples were analyzed by single-shot DIA-MS for cohorts Bleo (B) n = 5, bleomycin/TH5487 (BTH) n = 5, bleomycin/dexamethasone (DEX) n = 4, TH5487 only (TH) n = 4 and vehicle only (V) n = 4, respectively. Significance criteria; absolute fold-change ≥2 & FDR, Benjamini–Hochberg-corrected p-value ≤0.05 unless indicated otherwise. Source data are provided as a Source data file. Elements of this figure were created with BioRender.com.