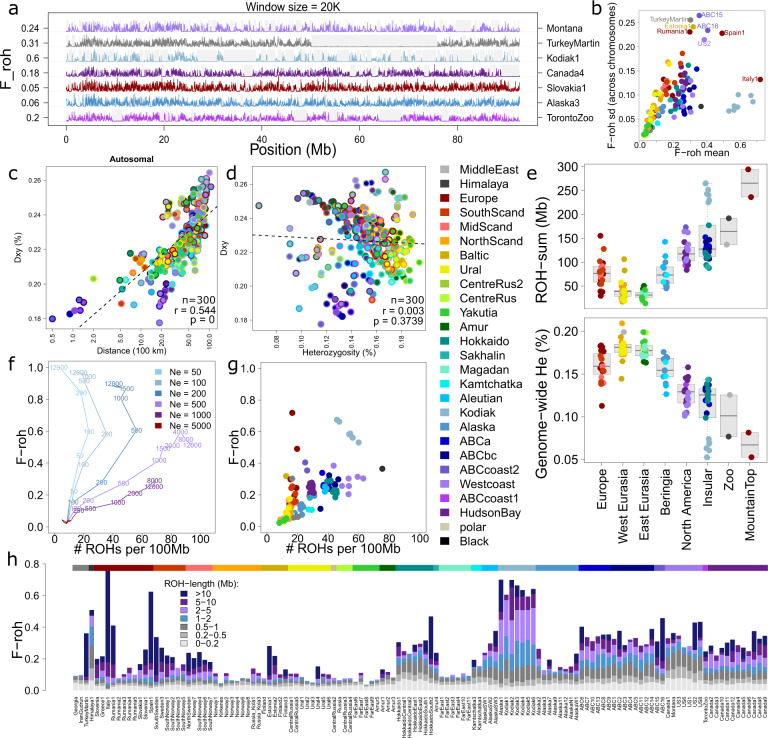
Fig. 6. Heterozygosity analyses reveal recent inbreeding on isolated mountain-tops, and ancient population bottlenecks in Kodiak and grizzly bears.
a Variation of heterozygosity levels (He) across a randomly chosen chromosome, for a random subset of samples, depicting He-levels of non-overlapping 20 kb windows, with grey segments highlighting the genomic regions which are marked as run of homozygosity (ROH) by the newly developed ROH-detection software Darwindow. Values along the y-axis represent sample and chromosome-specific FROH-values (proportion of chromosome marked as ROH). b Standard deviation of sample-specific FROH-values across chromosomes versus genome-wide mean FROH-values. High standard deviations are indicative of recent inbreeding events. c Mantel plot showing, for all (25 ∙ 24/2=) 300 brown bear population pairs, mean genome-wide genetic distance (DXY) versus geographic distance. d DXY-values against mean genome wide He-estimates, for all possible 300 population pairs. Population pairs involving bears from Middle East and North America affect the significance of the linear regression model. e Boxplots depicting sample-specific ROH-length and genome-wide He estimates, with samples grouped according to sample origin. Centre lines indicate median, and box limits indicate upper and lower quartiles. f SLIM2 genetic forward-in-time simulations indicate that following a population size reduction (from an initial Ne of 5000 individuals), and given sufficient time, ROH-distributions converge to a new equilibrium, with the relationship between FROH and number of ROHs depending on the new Ne. Shown are the number of generations since the population size reduction. g, h Sample specific ROH-distributions. The colour bar above the stacked barplot indicates population assignment.