Figure 1.
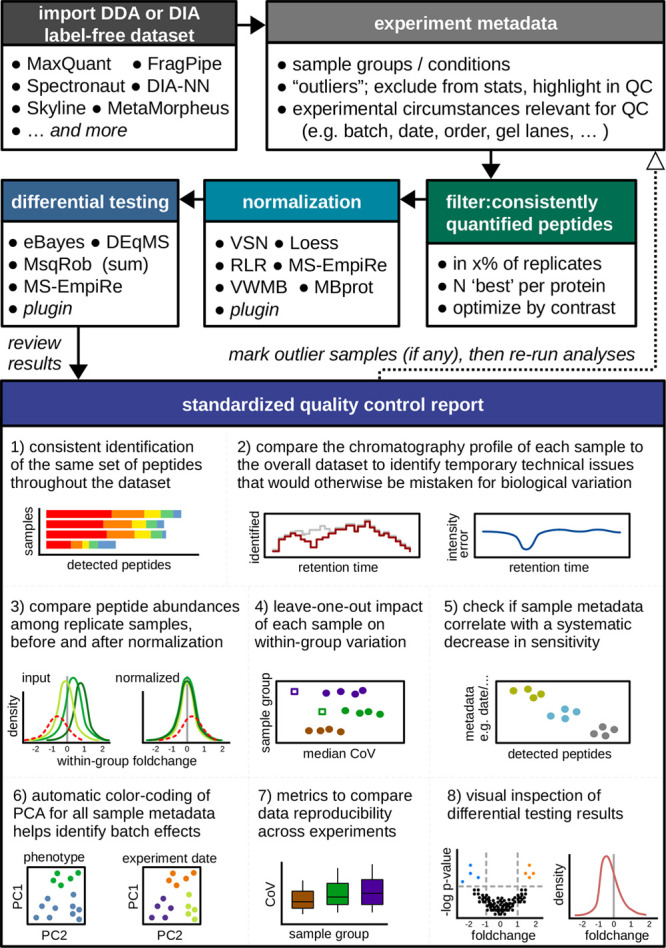
Overview of the MS-DAP workflow and data visualizations. MS-DAP is implemented as an open-source R package and provides a standardized solution for the analysis of label-free proteomics data sets generated from many software platforms. The pipeline facilitates multiple stages of quantitative data processing (filtering, normalization and stats) and conveniences application of pre-existing algorithms while also enabling bioinformaticians to provide custom functions (plugins) for normalization and differential testing. A comprehensive quality control report is generated that provides a multifaceted perspective on quality metrics, reproducibility, potential batch-effects and visualization of statistical results. User provided metadata describing experimental conditions, such as wet-lab sample handling steps or mass-spectrometry measurement order, are used to automatically generate respective quality control figures. The report is provided as a single PDF file that contains dozens of unique analyses, documentation thereof and information needed to reproduce results.
