Fig. 1 |. The commensal lifestyle of Staphylococcus epidermidis.
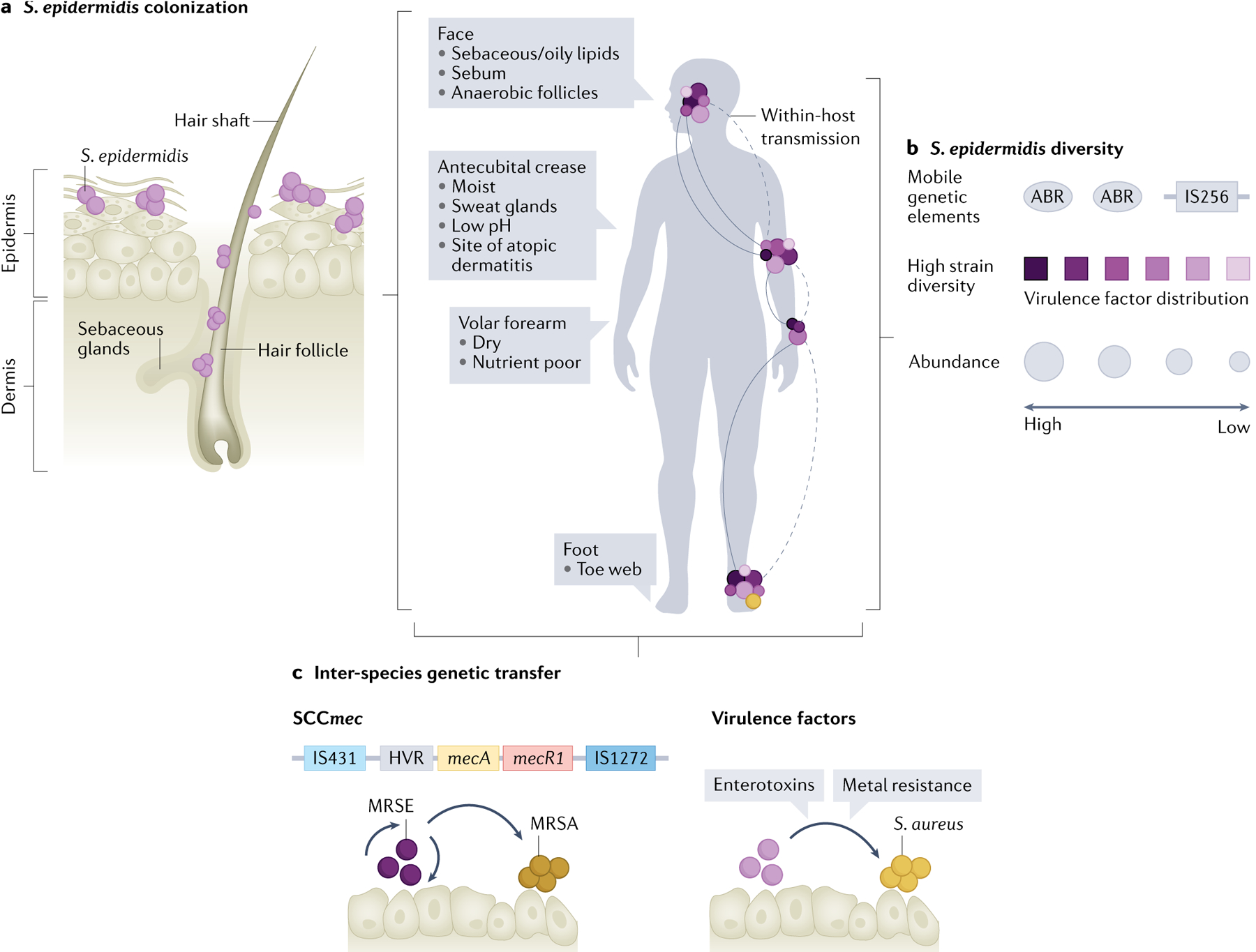
a | The skin can be subdivided into different ecological niches based on physio-chemical properties such as density of sebaceous and sweat glands, level of moisture and oxygen tension. Staphylococcus epidermidis colonizes all these environments at different densities, including the outermost stratum corneum as well as the interior of the hair follicle, and is a dominant colonizer of moist sites. b | Recent metagenomics studies revealed high strain-level heterogeneity of skin-colonizing S. epidermidis (represented by various colours) (BOX 1). These strains share genetic content, including virulence factors and antibiotic resistance factors (ABRs), via horizontal gene transfer of mobile genetic elements, including plasmids, phages and transposable elements (such as the insertion sequence IS256). Body sites are colonized by varying levels of S. epidermidis (indicated by circle size) with the highest density of S. epidermidis in moist or sebaceous sites. c | Horizontal gene transfer of the staphylococcal cassette chromosome mec (SCCmec), which encodes resistance to methicillin, between S. epidermidis and Staphylococcus aureus. Other virulence factors can also be exchanged between these organisms, including enterotoxins and genes for metal resistance. MRSA, methicillin-resistant S. aureus; MRSE, methicillin-resistant S. epidermidis.
