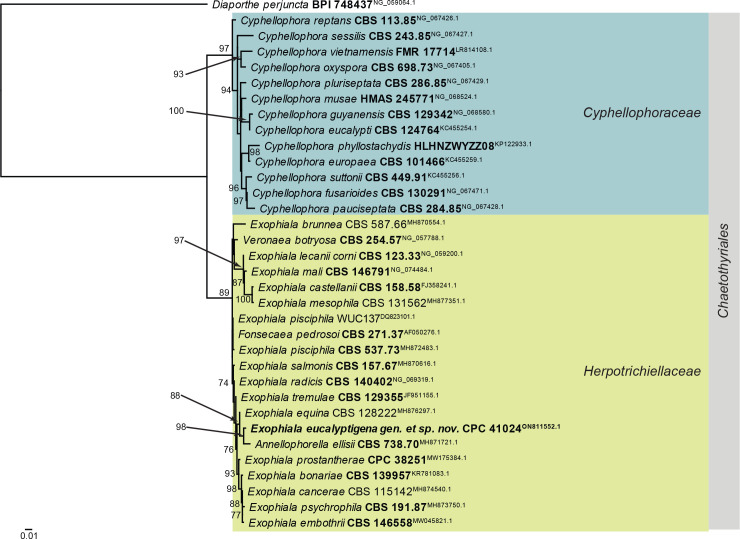
Fig. 3.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis of the Eurotiomycetes LSU nucleotide alignment. Bootstrap support values (> 69 % are shown; only values > 94 % are significant) from 5 000 ultrafast bootstrap replicates are shown at the nodes. Culture collection numbers and GenBank accession numbers (superscript) are indicated for all species. The tree was rooted to Diaporthe perjuncta (voucher BPI 748437; GenBank NG_059064.1) and the species treated here is highlighted with bold face. Accession numbers of sequence from material with a type status are also shown in bold face. Families and orders are indicated with coloured blocks to the right of the tree.