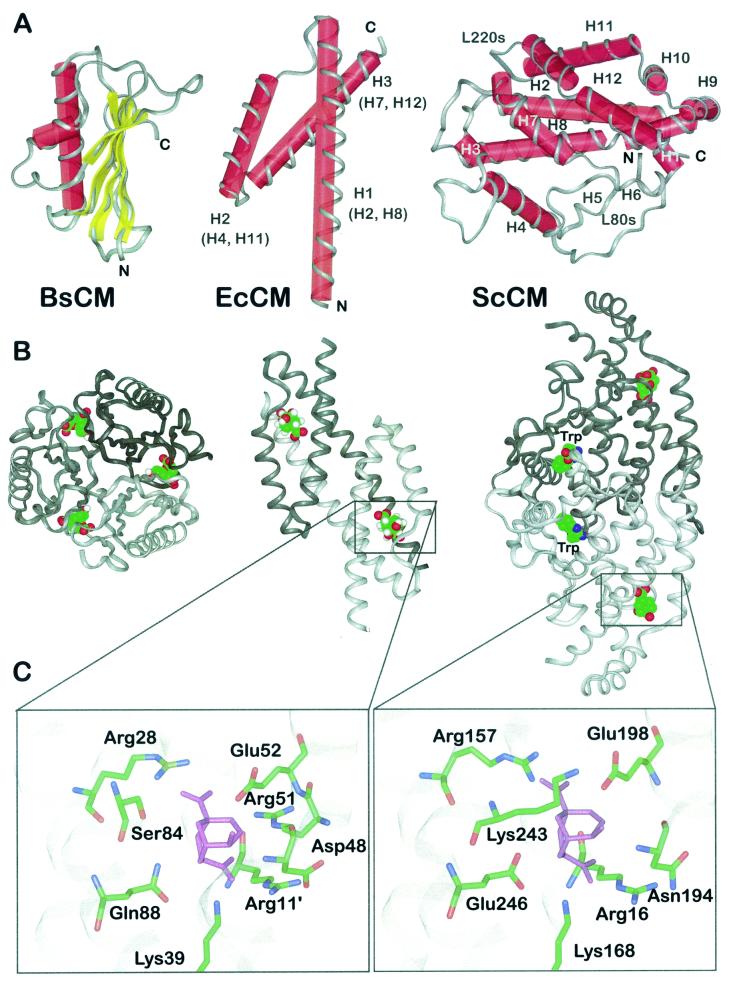
FIG. 3.
Structural prototypes of CM enzymes and binding mode of a stable transition state analogue. (A) Schematic presentations of the structural folds displayed by CMs from B. subtillis (BsCM) (left), E. coli (EcCM) (middle), and ScCM (right). The helix numbers in parentheses indicate the corresponding helices in the yeast enzyme. The polypeptide backbone is displayed in ribbon style, and secondary elements are labeled with red cylinders (α-helices) and yellow bars (β-sheets). N and C termini are indicated, as are structural elements of ScCM (see the text for details). (B) Oligomeric structure of B. subtilis CM (left), E. coli CM (middle), and ScCM (right) in complex with a stable transition state analogue. Monomeric subunits are indicated by different shades of grey. For ScCM, the binding position of the positive effector tryptophan is also shown. (C) Section views of the catalytic sites of E. coli CM (left) and ScCM (right) with the transition state analogue (purple) bound. Side chains are shown as sticks with atoms labeled by colour (green, carbon; blue, nitrogen; red, oxygen). Apostrophes indicate the position of the residue in an adjacent polypeptide chain.