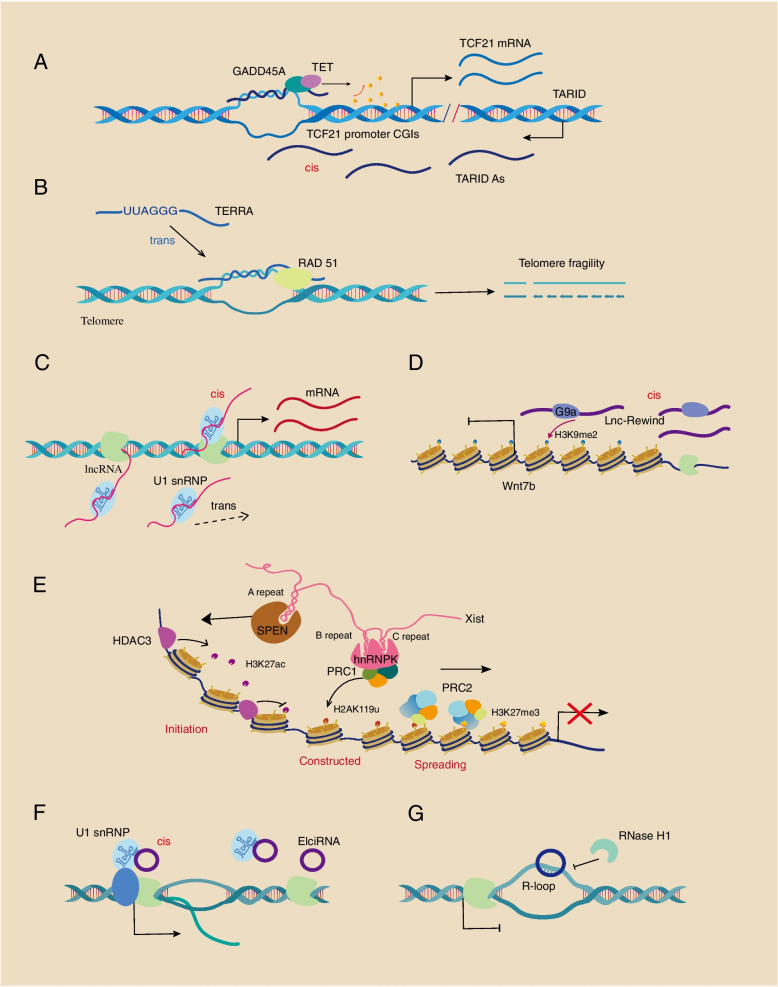
Fig. 2.
The mechanisms of chromatin-associated noncoding RNAs in gene regulation. A and B Chromatin-associated lncRNAs directly interact with DNA. A Antisense lncRNA TARID directly binds to the TCF21 promoter and is recognized by GADD45A and subsequent TET1, which results in DNA demethylation and TCF21 expression. B LncRNA TERRA with a specific sequence forms R-loops in telomeres and is recognized by RAD51, leading to telomere fragility. C-E Chromatin-associated lncRNAs indirectly interact with DNA or chromatin. C LncRNAs bind U1 snRNP to form a complex, which regulates transcription through RNA Pol II both in cis and trans. D LncRNA Rewind can recruit G9a to Wnt7b chromatin, leading to H3K9me2 deposition and gene repression. E Xist can bind many proteins, such as SPEN, hnRNP K, PRC1 and PRC2, to mediate transcriptional silencing, which is involved in the initiation, construction and spread of X-chromosome inactivation. F and G Chromatin-associated circRNAs directly or indirectly interact with DNA. F EIciRNAs within specific sequences interact with U1 snRNP at promoters and increase gene expression in cis. G CircRNA ci-ankrd52 forms R-loops by directly inserting DNA double strands, which will inhibit transcription, while RNase H1 can resolve R-loops and disarm the transcriptional inhibition effect. TCF21, transcription Factor 21; GADD45A, growth arrest and DNA damage inducible alpha; TET1, tet methylcytosine dioxygenase 1; TCF21, transcription factor 21; hnRNP K, heterogeneous nuclear ribonucleoprotein K