Fig. 1.
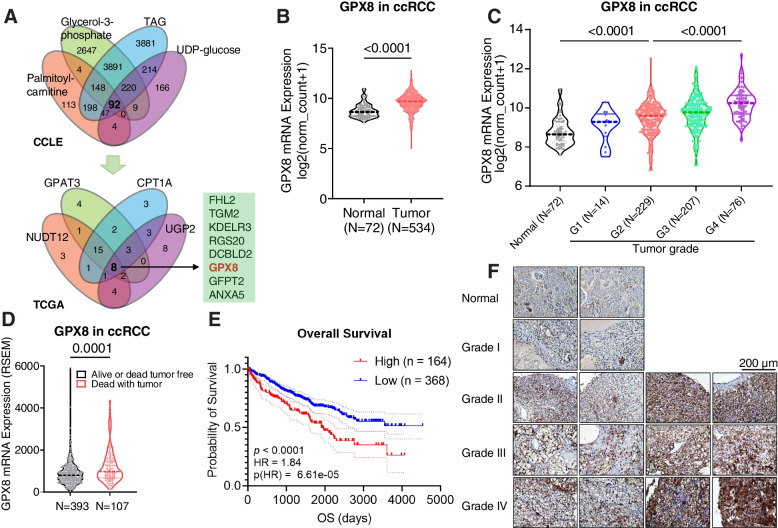
GPX8 is associated with higher grade and poor prognosis in ccRCC A, Two-tiered bioinformatic screening to find genes correlated with metabolites (top; CCLE database) and genes (bottom; TCGA-KIRC (Kidney renal clear cell carcinoma) database) of glycogen and lipid metabolism in ccRCC. The metabolites and genes used as queries are indicated in each Venn diagram. The criteria were set to FDR < 0.0001 and |r|< 0.15. The numbers in the Venn diagram represent the numbers of genes that meet the criteria. B-D, Violin plots for the mRNA expression levels of GPX8 according to (B) normal vs. tumor, (C) neoplasm histologic grade, and (D) alive or dead-tumor-free group vs. dead-with-tumor group among ccRCC patients. The mRNA expression values were obtained from the TCGA-KIRC dataset. P-values were determined by Mann–Whitney U test. E, Overall survival according to GPX8 mRNA expression for ccRCC patients from TCGA-KIRC database. F, GPX8 IHC staining for tumor array sections (KD482_biomax) from ccRCC patients. Magnification 400X