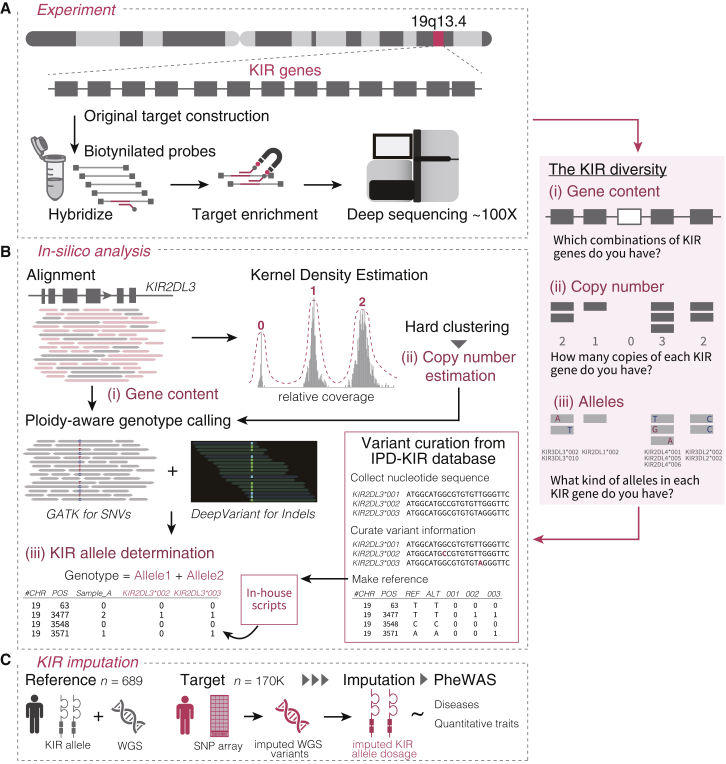
Figure 1.
Overview of this study
(A) Customized deep target sequencing of KIR genes was performed for 1,173 individuals.
(B) An integrative bioinformatics pipeline was devised to determine (i) the KIR gene content, (ii) copy number, and (iii) alleles. The coverage of aligned reads was used to determine the KIR gene content and copy number by kernel density estimation. The estimated copy number was used for ploidy-aware calling of SNVs and indels in the KIR region. Together with the public KIR allele database, the combination of SNVs and indels in the KIR region was used to determine the KIR alleles.
(C) With these KIR allele data and WGS data from 689 individuals, we implemented an in silico KIR imputation method. We imputed the KIR alleles in a large Japanese cohort (n = 169,907) to perform a PheWAS of the KIR alleles for 85 diverse complex traits.