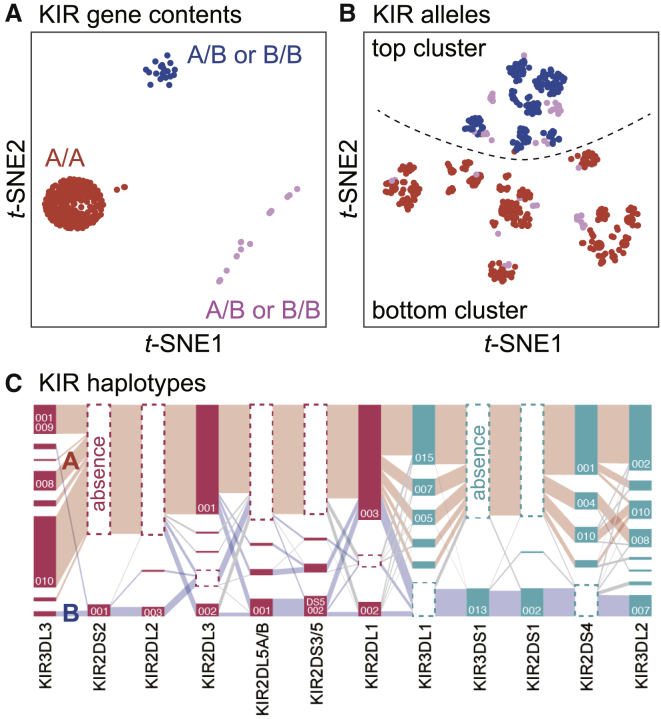
Figure 3.
The LD and haplotype structure of KIR region
(A) Unsupervised sample clustering by t-SNE based on KIR gene content. We manually categorized the plots into three clusters (red, blue, and pink). A/A, A/B, and B/B indicate the haplotype combination of the individuals in the cluster.
(B) Unsupervised sample clustering by t-SNE based on the KIR alleles. We annotated individuals with colors according to the clusters defined in (A). We segregated the points into top and bottom clusters by a dotted line.
(C) Visualization of haplotype structures of the KIR genes by Disentangler. The vertically stacked bars represent each of the KIR genes. A tile in a bar represents a KIR allele, and a segment connects two alleles on adjacent genes. Tiles with dotted frames indicate that the haplotype did not have the KIR gene. The height of each tile and the thickness of each segment correspond to the frequencies of the KIR allele and haplotype, respectively. The pink tiles are centromeric KIRs and the green tiles are telomeric KIRs. The representative haplotypes are colored manually in red (group A haplotypes) and in blue (group B haplotypes).