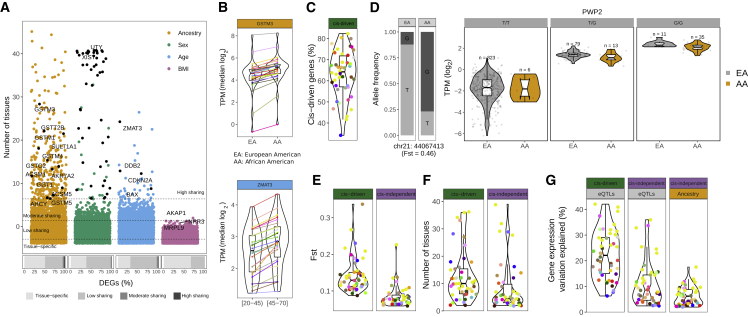
Figure 2.
Tissue sharing of DEGs and contribution of genetic variants to expression differences between human populations
(A) Distribution of the number of tissues in which a gene is DE with each demographic trait. Labeled ancestry- and age-DEGs correspond to highly tissue-shared genes enriched in glutathione-related metabolic processes and p53 pathways, respectively. For sex-DEGs, black points correspond to highly shared X-chromosome-inactivation (XCI) escapees17 and Y genes. As examples, the well-known XCI escapee XIST41 and the ubiquitously transcribed Y gene UTY are labeled. The top three most tissue-shared BMI-DEGs are labeled. Bottom bars show the proportions of tissue-specific DEGs and DEGs in a low (2–5), moderate (6–9), or high (≥10) number of tissues with each demographic trait.
(B) Median tissue expression values for a highly tissue-shared ancestry (top) and age (bottom) DEG (EA, European American; AA, African American).
(C) Percentage of cis-driven of the total ancestry eGenes DE across tissues.
(D) Example of a cis-driven DEG in sun-exposed skin. Left: bar plot showing the allele frequency of the eQTL variant in each population. Right: PWP2 violin plots of gene expression levels stratified by population and individual genotype.
(E) cis-driven DEGs are associated with eQTLs with larger Fst values (Wilcoxon signed-rank test, p = 1.9e−10). Violin plots show the distribution of tissue median Fst values for cis-driven (left) and cis-independent (right) DEGs.
(F) cis-driven DEGs are more tissue shared (Wilcoxon signed-rank test, p = 3.9e−07). Violin plots show the distribution of median tissue-sharing values for cis-driven (left) and cis-independent (right) DEGs.
(G) cis-eQTLs explain a larger amount of gene expression variation than ancestry (Wilcoxon signed-rank test, p = 2.8e−14). Violin plots show the distribution of tissue median gene expression variation explained by eQTLs in cis-driven and cis-independent DEGs (left) and by ancestry in cis-independent DEGs (right).