Figure 5.
Splicing patterns of ribosomal proteins vary across human populations
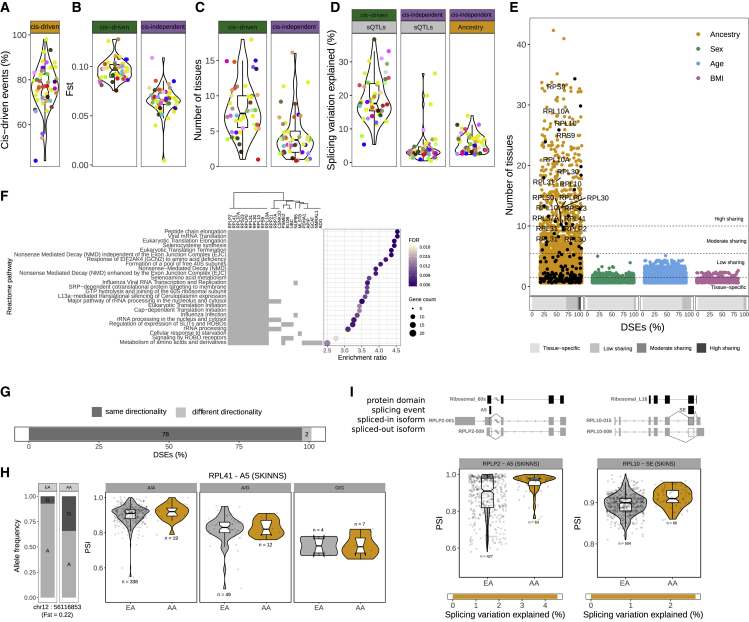
(A) Percentage of cis-driven DSEs across tissues.
(B) cis-driven DSEs are associated with sQTLs with larger Fst values (Wilcoxon signed-rank test, p = 1.563e−12). Violin plots show the distribution of tissue median Fst values for cis-driven (left) and cis-independent (right) DSEs.
(C) cis-driven DSEs are more tissue shared (Wilcoxon signed-rank test, p = 1.395e−06). Violin plots show the distribution of median tissue-sharing values for cis-driven (left) and cis-independent (right) DSEs.
(D) cis-sQTLs explain a larger amount of splicing variation than ancestry (Wilcoxon signed-rank test, p = 1.421e−13). Violin plots show the distribution of tissue median splicing variation explained by sQTLs in cis-driven and cis-independent DSEs (left) and by ancestry in cis-independent DSEs (right).
(E) Distribution of the number of tissues in which a splicing event is DS with each demographic trait. Ancestry-DSEs in ribosomal proteins are highlighted in black and labeled if shared in 10 or more tissues. Bottom bars show the proportions of tissue-specific DSEs and DSEs in a low (2–5), moderate (6–9), or high (≥10) number of tissues with each demographic trait.
(F) Functional enrichment of genes with highly shared ancestry-DSEs.
(G) Bar plot shows the proportion and number of ancestry-DSEs in ribosomal proteins in two or more tissues that have the same or different directionality.
(H) Example of a genetic variant associated with the splicing pattern of a ribosomal protein not previously reported as an sGene. Bar plot shows the allele frequencies in each population. Violin plots show the PSI distribution stratified by population and genotype. Points correspond to individual PSI values (EA, European American; AA, African American).
(I) Examples of two highly tissue-shared ancestry-DSEs on ribosomal proteins that affect a protein-coding domain.