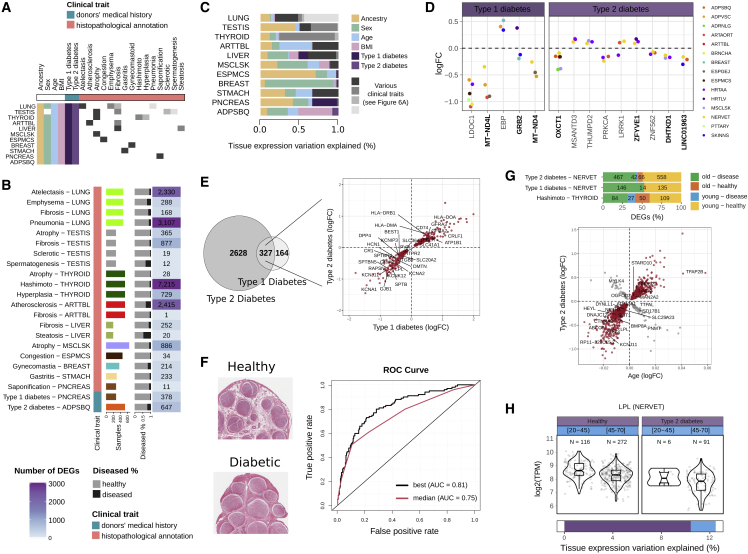
Figure 6.
Types 1 and 2 diabetes alter the transcriptome of multiple tissues, especially of the tibial nerve
(A) Clinical traits and affected tissues.
(B) Number of DEGs per tissue and clinical trait. Bars are colored according to the tissues.
(C) Proportion of the total tissue expression variation explained by each clinical trait.
(D) DEGs with type 1 or 2 diabetes in three or more tissues. The y axis corresponds to the fold change between healthy and diseased samples per tissue. In bold are known disease-related genes.70,71,72,73,74,75
(E) Left: the overlap between DEGs with types 1 and 2 diabetes in the tibial nerve. Right: DEGs with both diabetes have the same directionality. Genes driving functional enrichments (Table S6H) are labeled in the plot.
(F) On the left, tissue images from a healthy and a diabetic individual. Note the larger diameter of the fascicles (circles) and smaller interstitial spaces in the diabetic donor, consistent with previous observations.76 The right shows the ROC curves of the top and a median performing classifier.
(G) Top: DEGs with age and clinical traits show a biased directionality. Our observations are not confounded by age differences between healthy and diseased individuals (Figure S7F). Bars indicate the proportion (and number) of DEGs in each of the four possible directionalities. Bottom: scatterplot shows the fold change associated with age (x axis) versus the fold change between healthy and type 2 diabetes (y axis) in the tibial nerve. Labeled genes (24) have been previously associated with type 2 diabetes susceptibility in the tibial nerve through transcriptome-wide association studies.77
(H) LPL expression changes with age and type 2 diabetes. Gene expression levels are represented as boxplots with samples stratified by both traits. Bars at the bottom indicate the proportions of expression variation explained by age and type 2 diabetes.