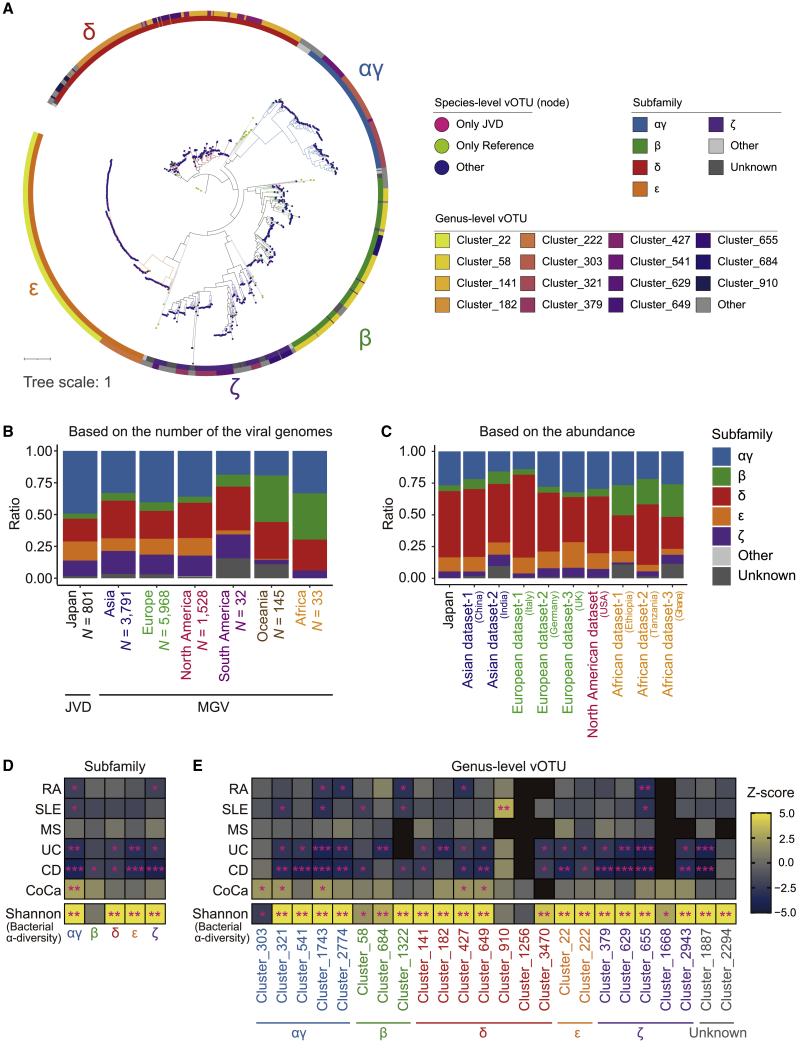
Figure 5.
Interpopulational and case-control comparisons of the crAss-like phages
(A) A maximum-likelihood phylogenetic tree reconstructed from the TerL proteins of the crAss-like phages. The color of the nodes represents the species-level clusters present only in the JVD (magenta), present only in the reference (green), or neither of the cases (navy). The outer rings represent the subfamily-level (inner) and genus-level (outer) taxonomic annotation of the crAss-like phages.
(B) A bar plot depicting the compositions of the subfamilies of the crAss-like phage genomes for the JVD and MGV. The genomes from the MGV are grouped according to their continental origin.
(C) A bar plot depicting the compositions of the subfamilies of the crAss-like phages calculated from the abundances (RPKM) in each group.
(D and E) Heatmaps represent the association of the crAss-like phages to the diseases (upper) and Shannon index (lower) at the subfamily (D) and genus (E) level, respectively. The colors indicate the Z score in each test. ∗p < 0.05. ∗∗p < 0.05/number of clades (per objective variables). ∗∗∗p < 0.05/number of tests across all diseases (only for association tests for diseases). See also Tables S8, S9, S10, and S11.