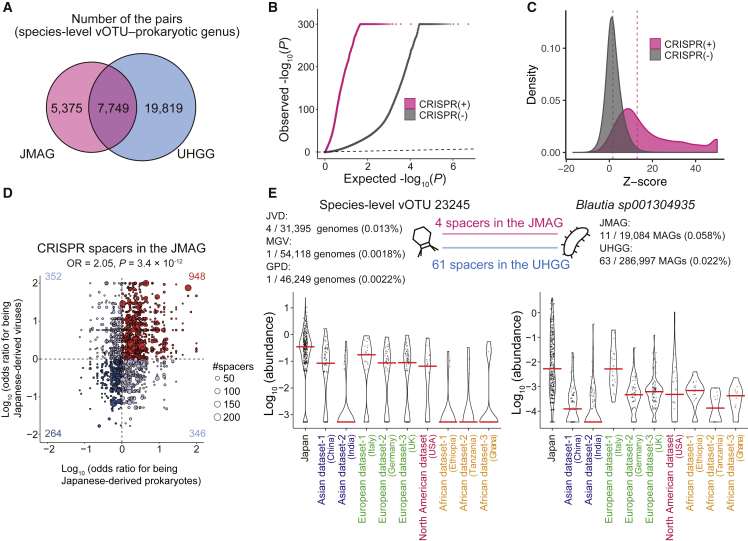
Figure 6.
Virus-prokaryote interaction analysis based on the CRISPR and abundances
(A) Number of the pairs of the species-level vOTU and prokaryotic genus detected in the JMAG and UHGG.
(B) A quantile-quantile plot of the p values from the virus-prokaryote association analysis stratified by whether the virus-prokaryote pairs are supported by the CRISPR spacers in the JMAG (magenta) or not (gray). The x axis indicates −log10(P) expected from the uniform distribution. The y axis indicates the observed −log10(P). The diagonal dashed line represents y = x, which corresponds to the null hypothesis.
(C) A density plot of the Z score from the virus-prokaryote association analysis stratified by whether the virus-prokaryote pairs are supported by the CRISPR spacers in the JMAG (magenta) or not (gray). The upper limit of the Z score is set at 50. The diagonal dashed line represents y = x, which corresponds to the null hypothesis. The vertical dashed lines indicate the mean of the Z score for each group of the virus-prokaryote pair.
(D) A scatterplot of the odds ratios for being the Japanese-derived viruses (y axis) and prokaryotes (x axis) for the virus-prokaryote pairs supported by the CRISPR in the JMAG. The size of the dots represents the number of spacers supporting the virus-prokaryote pairs. The horizontal and vertical dashed lines represent odds ratio = 0 for virus and prokaryote, respectively.
(E) Violin plots of the species-level vOTU 23245 (left) and Blautia sp001304935 (right) abundances (RPKM) in each group. The red center lines indicate the median values. See also Figure S10 and Tables S12 and S13.