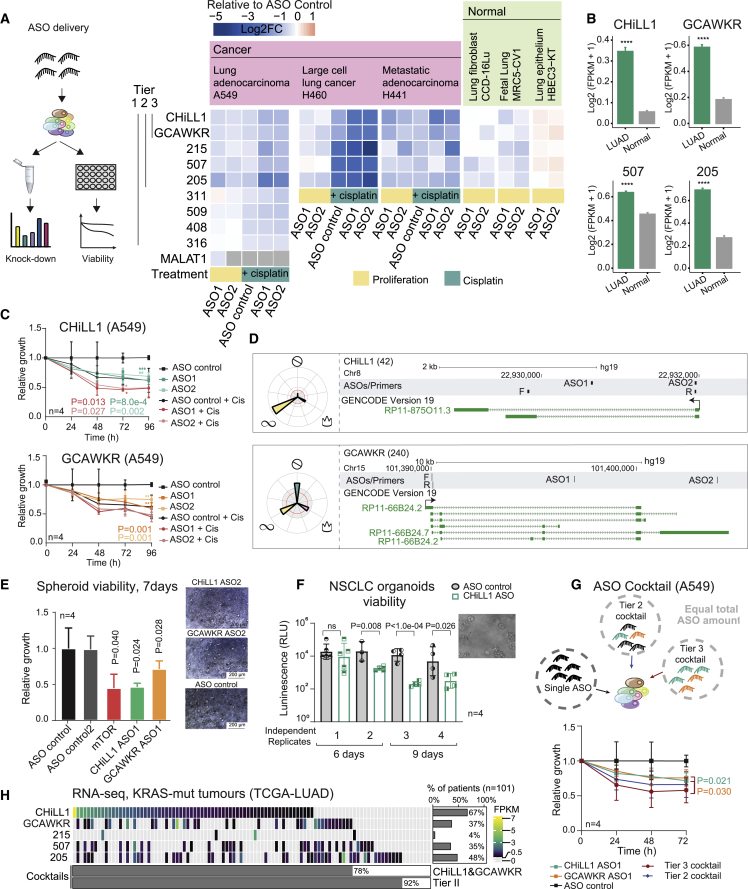
Figure 5.
Therapeutic targeting of oncogenic lncRNAs CHiLL1 and GCAWKR
(A) (Left) Experimental workflow to test knockdown efficiency and phenotypic effect of ASO transfection. (Right) Summary of all ASO/cell line results. Rows: targeted lncRNAs; columns: ASOs and cell lines. Values reflect the mean log2 fold change in viability following ASO transfection and are normalized to the control non-targeting ASO for proliferation, n > 2. Numbers to the left indicate library candidate identifiers and are grouped into tiers. Each lncRNA is targeted by two independent ASO sequences (1 and 2, below). MALAT1 lncRNA is used as positive control whose knockdown is expected to decrease viability. “Normal” cells are non-transformed cells of lung origin.
(B) Expression of tier 2 lncRNAs—CHiLL1, GCAWKR, candidate 205 (LINC00115), candidate 507 (LINC00910)—in TCGA RNA-seq. LUAD: 513 samples; normal: 59 samples. Statistical significance estimated by Student's t test; ∗∗∗∗p < 0.0001. Data were not available for candidate 215 in the TCGA dataset.
(C) A549 cell growth upon transfection with two independent ASOs. Results are normalized to non-targeting control ASO (n = 4 biological replicates; error bars: SD statistical significance: two-tailed Student’s t test).
(D) Genomic loci encoding CHiLL1 (top panel) and GCAWKR (bottom panel).
(E) (Left) Viability of H441 spheroid cultures 7 days after ASO transfection (25 nM). mTOR ASO was used as a positive control (n = 4 biological replicates; error bars: SD; one-tailed Student’s t test). (Right) Representative images (Leica DM IL LED Tissue Culture Microscope).
(F) Viability of independent replicates of BE874 organoids grown from a single KRAS+ patient-derived xenograft after CHiLL1 ASO transfection. The error bars represent the variability of technical replicates, while the biological replicates are indicated on the bottom (n = 4 biological replicates; n > 3 technical replicates; error bars: SD; one-tailed Student’s t test).
(G) ASO cocktails. (Top) For all experiments, the total amount of ASO did not vary (25 nM). Cocktails were composed of equal proportions of indicated ASOs. (Bottom) A549 cell populations, normalized to non-targeting ASO (n = 4 biological replicates; error bars: SD; statistical significance: one-tailed Mann-Whitney test).
(H) Expression of Tier 2 lncRNAs in KRAS+ LUAD tumors (TCGA, n = 101). Each cell represents a patient and is colored to reflect expression as estimated by RNA-seq (expression defined as >0.5 FPKM). At the bottom is the percentage of patients with at least one lncRNA from the indicated cocktails.