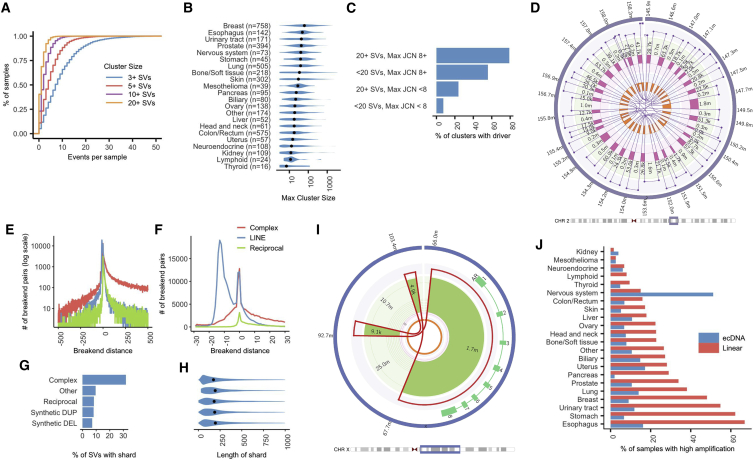
Figure 4.
Complex rearrangements and high amplification
(A) Cumulative distribution function plot of count of complex rearrangement clusters per sample with at least 3, 5, 10, and 20 variants.
(B) Violin plot showing the distribution of the maximum number of variants in a single complex rearrangement cluster per sample, grouped by tumor type. Black dots indicate the median values for each tumor type.
(C) Proportion of clusters contributing to at least one amplification, deletion, homozygous disruption, or LOH driver in a panel of cancer genes by complexity of cluster and maximum JCN.
(D) Fully resolved chromothripsis event consisting of 31 structural variants affecting a 13-Mb region of chromosome 2 in HMF001571A, a prostate tumor.
(E) Counts of occurrences of trans-phased breakends by distance between the breakends for complex events, LINE insertions, and two-break reciprocal clusters in the range of −500 to 500 bases (log scale). Negative distances indicate overlapping breakends and duplication at the rearrangement site.
(F) Counts of occurrences of trans-phased breakends by distance between the breakends zoomed in to −30 to 30 bases.
(G) Proportion of variants with at least one breakend joining a shard of less than 1 kb in length by resolved type for selected resolved types.
(H) Violin plot showing the distribution of shard length by resolved type.
(I) Double minute formed by three junctions in HMF003969A, a prostate tumor, and which amplifies known oncogene, AR, to a copy number of approximately 23.
(J) Proportion of samples with ecDNA and linear amplifications by cancer type.
See also Figures S6 and S7 and Table S5.