Figure 5.
Clinically relevant rearrangements
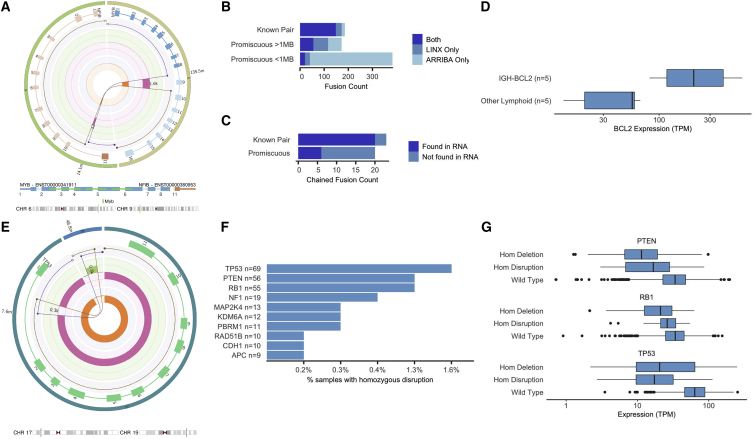
(A) A MYB-NFIB fusion caused by a reciprocal translocation in HMF000780A, a salivary gland tumor. The translocation links exons 1–8 in MYB to exon 11 in NFIB.
(B) Comparison of LINX fusion predictions in Hartwig cohort to Arriba fusion predictions from orthogonal RNA sequencing for known pairs and promiscuous fusion partners. Promiscuous fusions of less than 1 Mb length are shown separately, as they may occur from readthrough transcripts and not be associated with a genomic rearrangement.
(C) Count of LINX chained fusion predictions for known and promiscuous fusion and whether they are also found to be expressed in RNA by Arriba.
(D) Distribution of BCL2 expression in lymphoid samples with and without a predicted pathogenic IGH-BCL2 rearrangement. Box: 25th–75th percentile; whiskers: data within 1.5 times the interquartile range (IQR).
(E) Reciprocal translocation affecting TP53 in HMF001913A, a prostate tumor. The two predicted derivative chromosomes overlap by approximately 300 bases on both ends but are trans phased, which rules out the possibility of a templated insertion at either location. Although the TP53 copy number alternates between one and two, no derivative chromosome contains the full gene and the gene is homozygously disrupted.
(F) Prevalence of homozygous disruption drivers for top 10 most affected tumor-suppressor genes.
(G) Distribution of gene expression in Hartwig cohort for samples with homozygous deletion, homozygous disruption, and wild type for each of RB1, TP53, and PTEN. box: 25th–75th percentile; whiskers: data within 1.5 times the IQR.