Figure 1.
Visualization of mRNA expression across FFPE tissue sections with adapted protocol
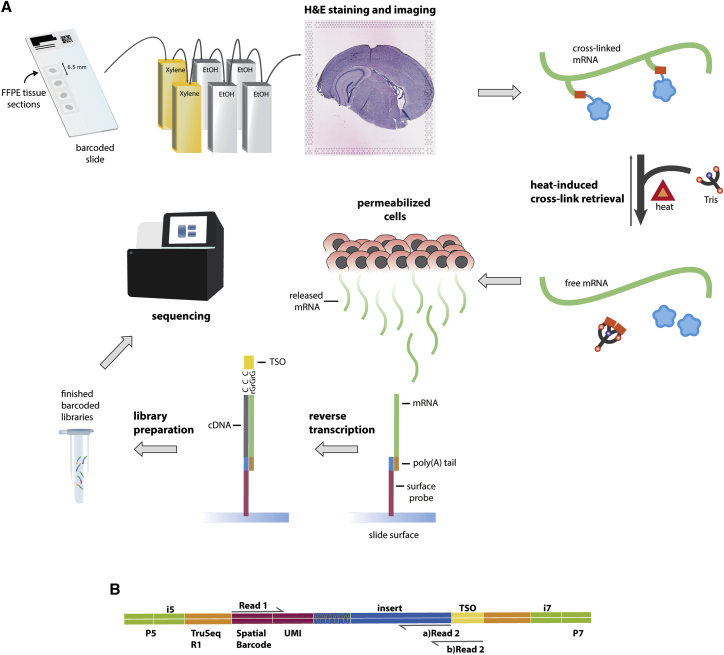
(A) Tissue sections are placed on spatial gene expression slides. The slides are deparaffinized following a standard protocol (see STAR Methods). H&E-stained sections are imaged under a high-resolution microscope. Cross-links are reversed by heat and in the presence of Tris-EDTA buffer at pH 8.0. Tissues are permeabilized so that mRNA diffuses to the barcoded surface probes and hybridizes with its oligo(dT) capture region. A cDNA strand is synthesized by reverse transcription. Finished libraries are sequenced either with custom primer for read 2 reverse complementary to template switch oligo (TSO) sequence or with conventional Truseq R2 primer.
(B) Final construct overview. Read 2 can begin in 2 different positions depending on whether the sequencing has been performed utilizing a custom primer (see STAR Methods).