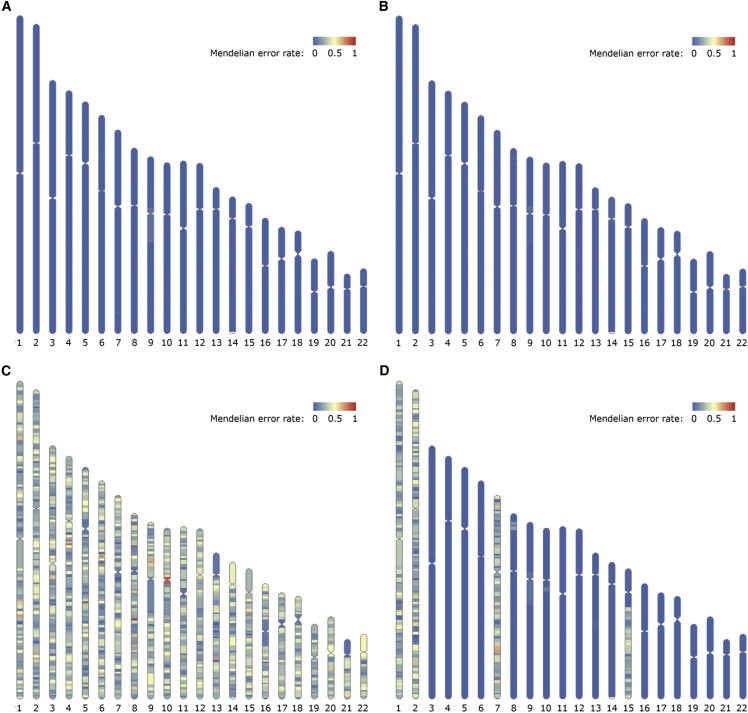
Figure 4.
Mendelian error rates show that PofO phasing correctly infers parental haplotypes
(A) The inferred maternal haplotype for HG005 (child) is compared with the ground-truth variant genotypes for HG007 (mother). The Mendelian error rate is low across all chromosomes.
(B) The inferred paternal haplotype for HG005 (child) is compared with the ground-truth variant genotypes for HG006 (father).
(C) The inferred maternal haplotype for HG005 (child) is compared with the ground-truth variant genotypes for HG006 (father). This is the expected pattern if a PofO is misassigned for all chromosomes.
(D) We artificially produced chromosomes and regions with incorrect PofO assignments in the comparison of HG005’s maternal haplotype with HG007. The lack of such regions is evidence that the PofO phasing method correctly distinguishes maternal and paternal homologs. We switched the maternal and paternal haplotypes for chromosomes 1, 2, and 7 to simulate erroneous iDMR inferences, and we created a large switch error on chromosome 15 by reducing the bin size in the BreakpointR step.33
See also Figures S13–S16.