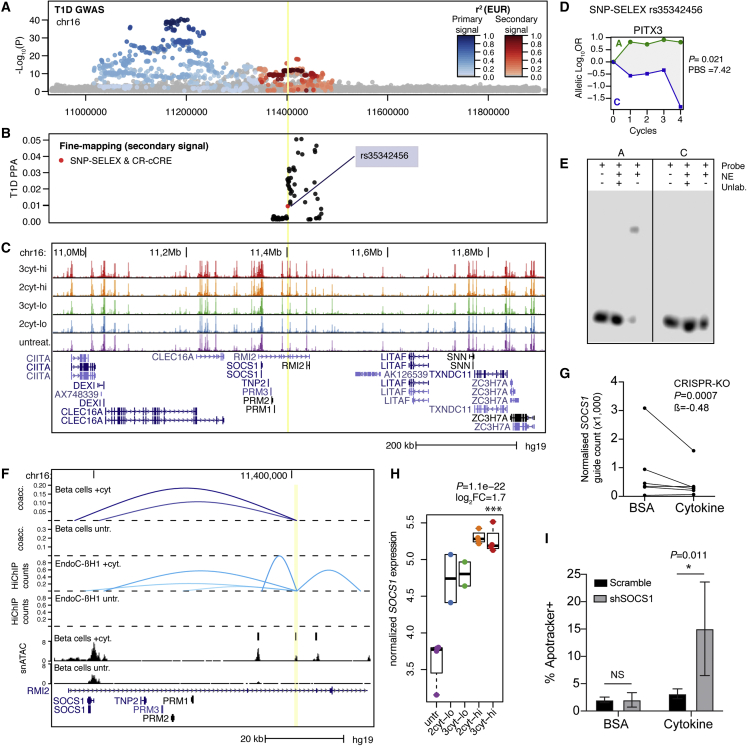
Figure 6.
T1D locus 16p13 regulates beta cell survival gene SOCS1 in cytokine exposure
(A) Regional plot showing T1D association for two independent signals at the DEXI/SOCS1 locus.
(B) Fine mapping probabilities of the secondary signal. Variants with SNP-SELEX effect on differential TF binding and within a cytokine-responsive cCRE are highlighted in red.
(C) Genome browser of the locus showing bulk ATAC-seq. Treatment abbreviations as in Figure 1.
(D) SNP-SELEX results for variant rs35342456.
(E) EMSA with nuclear extract (NE) from MIN6 cells showing preferential binding to the reference allele.
(F) Zoom in of the locus showing location of variant rs35342456 (yellow line) in a cytokine-responsive beta cell cCRE with co-accessibility and 3D interaction to the SOCS1 promoter in cytokine-treated and untreated EndoC-βH1 cells. For 3D interactions, virtual 4C counts are scaled between 0 and 1.
(G) Counts of each sgRNA in the CRISPR-KO screen targeting SOCS1 in untreated and high-dose cytokine EndoC-βH1, normalized to the sequencing depth of each sample. Effect size and uncorrected p value from MAGeCK.
(H) Normalized expression of SOCS1 in human islet samples in cytokines. Log2 fold change and uncorrected p values from DESeq2 comparing high-dose three-cytokine (red) and untreated (purple).
(I) Quantification of apoptotic EndoC-βH1 cells with shRNA targeting SOCS1 or scramble in either high-dose cytokine or vehicle (0.1% BSA). Values are mean and error bars are SD from n = 4 transductions, and p values are shown from two-way ANOVA followed by pairwise comparisons using Tukey’s HSD.