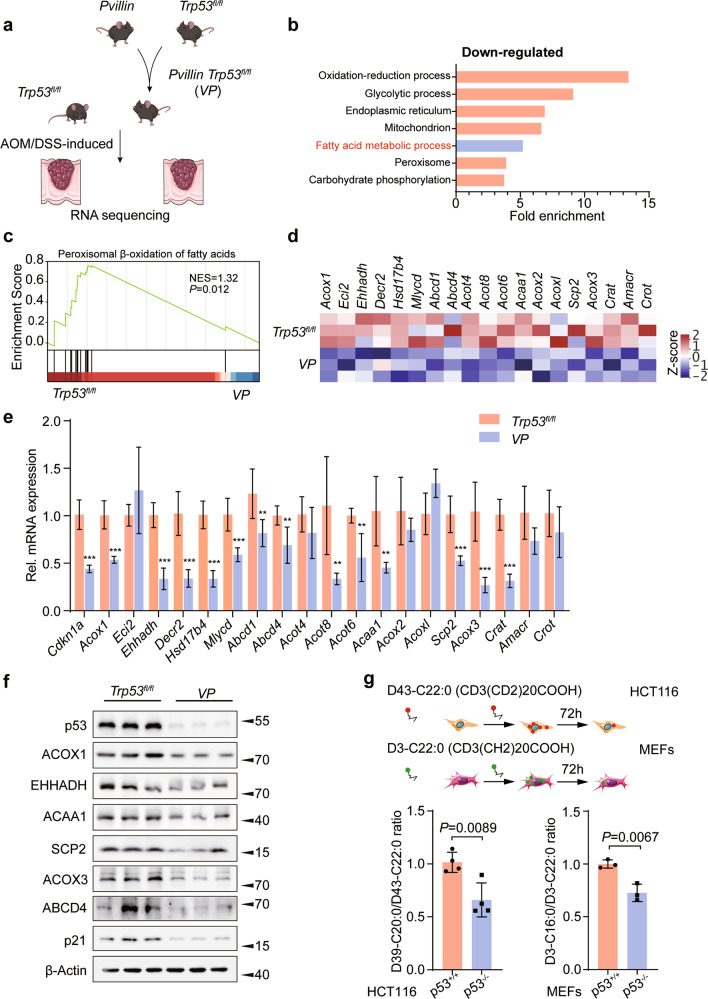
Fig. 1. p53 facilitates peroxisomal β-oxidation.
a Experimental scheme. Trp53fl/fl mice were crossed with Pvillin-Cre mice to generate Pvillin-Cre + Trp53fl/fl (VP) mice, and the colorectal tumor model was induced by AOM/DSS. b Gene ontology analysis of gene sets that were significantly downregulated in metabolic process from VP mice CRC tissues compared to Trp53fl/fl ones. c Gene set enrichment analysis (GSEA) was used to evaluate changes in the gene signature of peroxisomal FAO comparing CRC tumors from Trp53fl/fl and VP mice. d Heatmap of RNA-seq expression values of peroxisomal FAO genes comparing tumors from VP and Trp53fl/fl mice. e qRT-PCR analysis of various peroxisomal FAO genes comparing CRC tumors from Trp53fl/fl and VP mice, n = 6. f Western blot analysis for ACOX1, EHHADH, ACAA1, SCP2, ACOX3, ABCD4, and p21 expression in CRC tumors from AOM/DSS-induced colitis-associated colorectal cancer (CAC) mouse model, n = 3. g HCT116 cells and MEFs were incubated with D43-C22:0 or D3-C22:0, whose catabolism to D39-C20:0 or D3-C16:0 was measured using mass spectrometry. Peroxisomal FAO was expressed as the ratio of D39-C20:0 to D43-C22:0 or D3-C16:0 to D3-C22:0; n = 3 or 4. Data were presented as mean ± SD. **P < 0.01, ***P < 0.001.