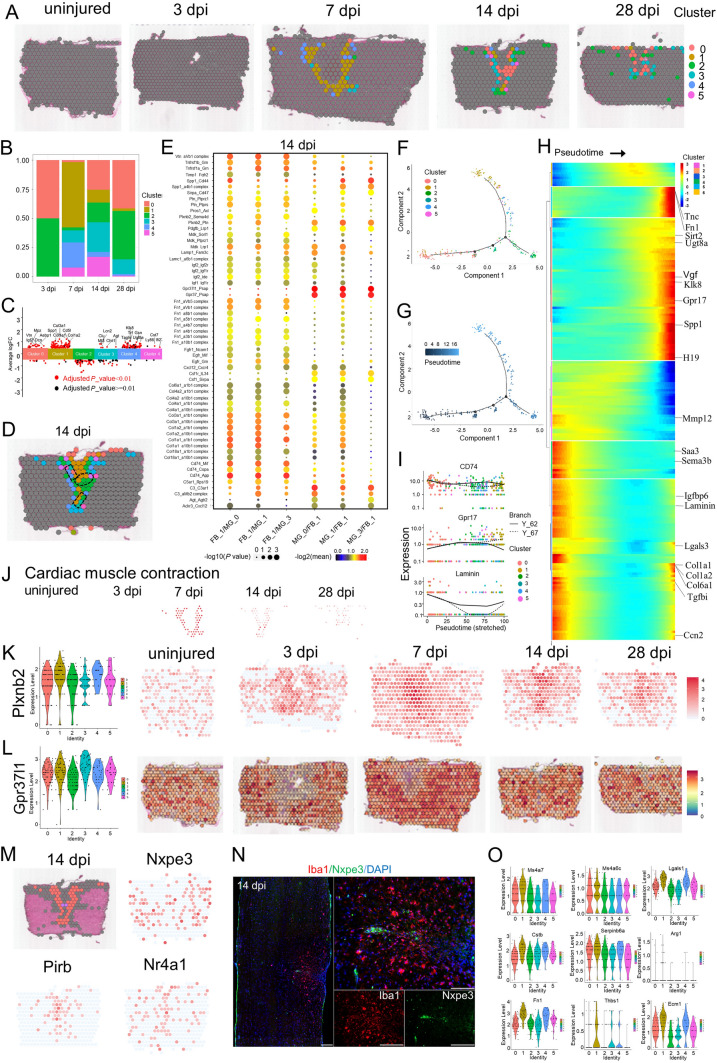
Fig. 6.
Phenotypic and functional heterogeneity of microglia in the glial scar. A Spatial maps showing the distribution of 6 microglia clusters in the glial scar at different time points after injury. B Bar plot showing the fraction of all spots comprising each subpopulation at different time points after injury. C Jitterplot showing the top 5 differentially-expressed genes (DEGs) in each subpopulation. D Definition of the boundary areas to study the interaction between two neighboring microglia clusters in the scar. Regions 2 spots wide along the boundary lines in each cluster are selected. E Dot plot showing the mean interaction scores between neighboring clusters at the boundaries for ligand-receptor pairs. The ligand-receptor pairs are listed on the left. The size of a circle denotes the p-value, and the color denotes the mean interaction score. F, G Trajectory of microglia in the glial scar. H Heat map showing the changes in genes expression along a spatial trajectory. I Correlation of the expression of CD74, Gpr17, and laminin with pseudotime. J Spatial maps showing the GSVA score of the cardiac muscle contraction pathway enrichment in cluster 1 cells at different time points after injury. K Violin plots and spatial maps showing the expression of Plxnb2 at different time points after injury. L Violin plots and spatial maps showing the expression of Gpr37l1 at different time points after injury. M Spatial maps showing the expression of selected DEGs in spots that cross the fibroblast scar and form bridges at 14 dpi. N IF-stained 14-dpi scars for Nxpe3 (red) and DAPI (blue) (scale bars, 200 µm). O Violin plots showing the expression of selected genes contributing to microglia-organized scar-free spinal cord repair in neonatal mice.