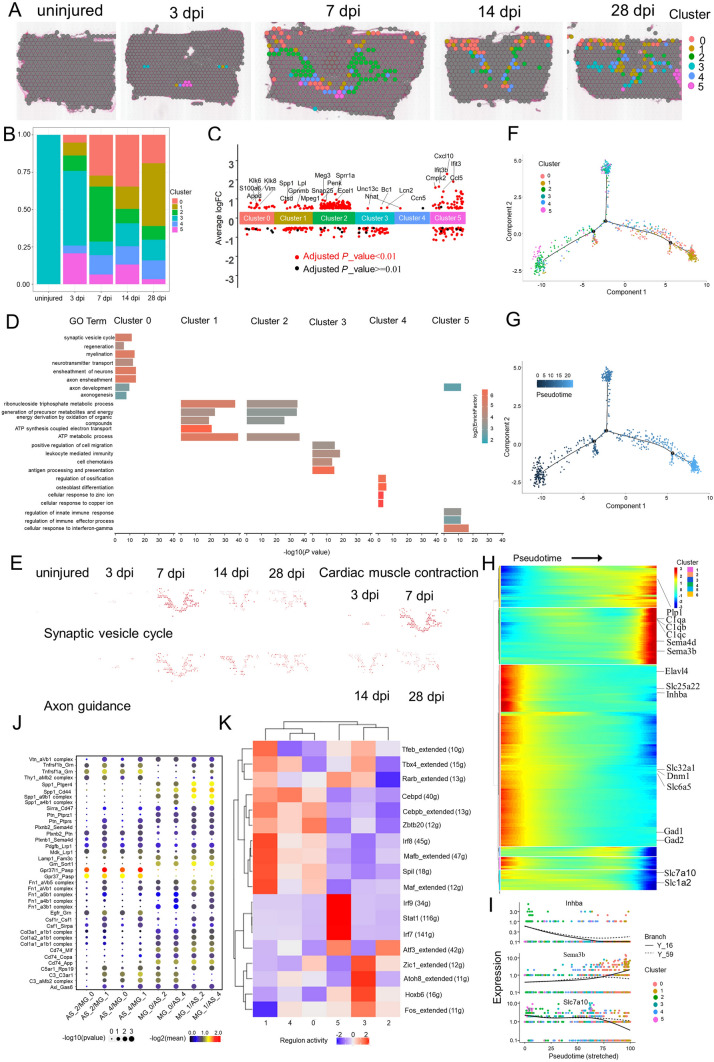
Fig. 7.
Phenotypic and functional heterogeneity of astrocytes in the glial scar. A Spatial maps showing the distribution of 6 astrocyte clusters in the glial scar at different time points after injury. B Bar plot showing the fraction of all spots comprising each subpopulation at different time points after injury. C Jitterplot showing the top 5 differentially expressed genes in each subpopulation. D Selected GO terms that were enriched for each cluster using Fisher’s exact test. E Spatial maps showing the GSVA score of the synaptic vesicle cycle and axon guidance pathways enriched in cluster 0 cells, and the cardiac muscle contraction pathway enriched in cluster 1 cells at different time points after injury. F, G Trajectory of astrocyte in the glial scar. H Heat map showing the changes in gene expression along a spatial trajectory. I Expression of Inhba, Sema3b, and Slc7a10 correlates with pseudotime. J Dot plot showing the mean interaction scores between neighboring clusters at the boundaries for ligand-receptor pairs. The ligand-receptor pairs are listed on the left. The size of a circle denotes the p-value and the color denotes the mean interaction score. K Heat map showing the enriched transcription factors by single-cell regulatory network inference and clustering analysis in different clusters after injury.