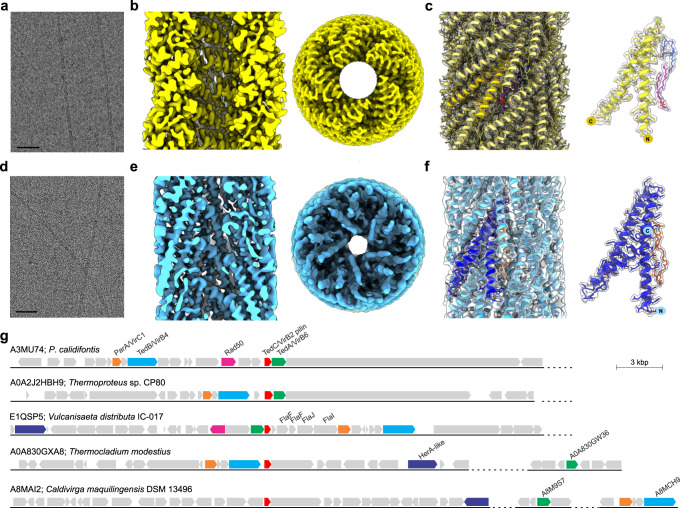
Fig. 1. Archaeal conjugative pili.
a, d Representative cryo-electron micrographs of A. pernix and P. calidifontis pili, respectively, from >8000 images collected for each. Scale bars, 50 nm. b, e Side and top views of the A. pernix (b, yellow) and P. calidifontis (e, blue) cryo-EM density maps at resolutions of 3.3 Å and 4.0 Å, respectively. The front half of the filament has been removed in the side views, so that lumens are visible. c, f Atomic models in ribbon representation of the A. pernix (c, yellow) and P. calidifontis (f, blue) pili docked within their respective transparent cryo-EM density maps. The asymmetric unit of the A. pernix pilin CedA1 (c, bright yellow) shows two bound lipids (magenta and blue), while a single asymmetric unit of the P. calidifontis pilin TedC (f, dark blue) shows one bound tetraether lipid (orange). g Genomic loci encompassing the Ted system in different members of the order Thermoproteales. Each of the five genera within the Thermoproteales (genera Caldivirga, Pyrobaculum, Thermocladium, Thermoproteus, and Vulcanisaeta) is represented. Genes encoding the conserved VirB2/CedA1-like pilin protein TedC, VirB6/CedA-like transmembrane channel TedA, and VirB4/CedB-like AAA + ATPase TedB are shown as red, green, and cyan arrows, respectively. Additional conserved genes are also indicated, including VirC1/ParA-like, Rad50, and HerA-like helicase which are shown as orange, magenta, and dark blue arrows, respectively. Other genes are shown in gray. Genomic loci are aligned using the TedC gene and indicated with the corresponding UniProt accession numbers, followed by the organism name. In some species, the components of the Ted systems are encoded within distal genomic loci which are separated from the TedC-encoding loci by dashed lines, and the corresponding genes are identified with their UniProt accession numbers.