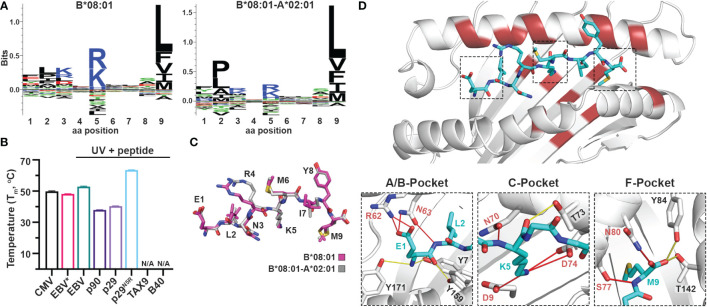
Figure 4.
Introduction of a P5 anchoring specificity into the C-pocket of HLA-A*02:01 using a B*08:01 structural template. (A) The sequence logos of the HLA-B*08:01 (left), and B*08:01-A*02:01 (right) rendered using an in-house method and visualized in Seq2Logo from the NetMHCpan4.0 (40). (B) Thermal stabilities of HLA-B*08:01-A*02:01 refolded with CMV (ELNRKMIYM) or EBV* (FLRGRAJGL, where J is the 3-amino-3-(2-nitrophenyl)-propionic acid) and after UV-irradiation in the presence of 10-fold molar excess of EBV, p90, p29, p29N5R, TAX9, and B40 peptides. Data are mean ± SD obtained from n = 3 technical replicates. N/A, no exchange. (C) Overlay of the CMV peptide bound to the chimeric B*08:01-A*02:01 (grey) and wild-type B*08:01 (magenta) molecules. (D) Crystal structure of HLA-B*08:01-A*02:01/CMV complex where substitutions of the groove residues are highlighted in red. Hydrogen bonds and salt bridges between the peptide and the base or groove allele residues are shown as yellow or red lines, respectively. Peptide, A*02:01-specific, and B*08:01-specific residues are labeled in cyan, black, and red font, respectively.