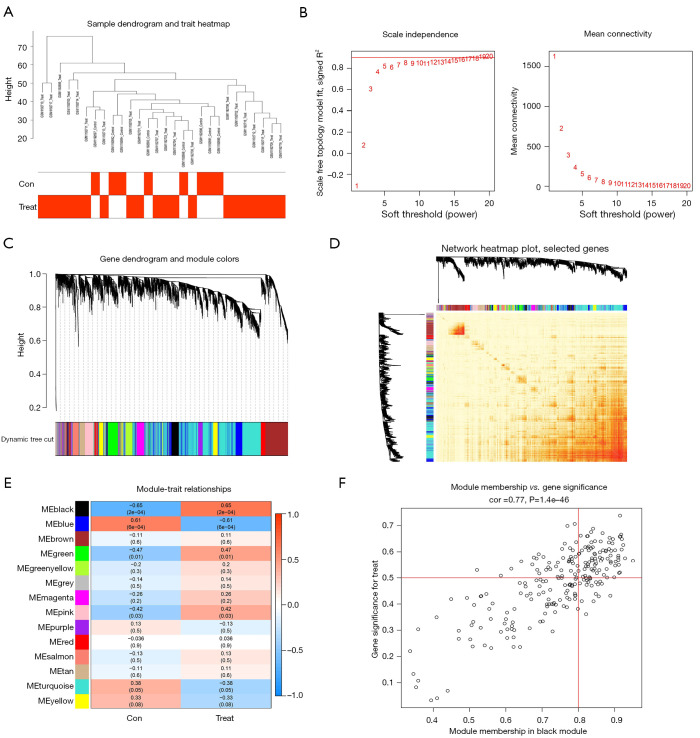
Figure 11.
WGCNA selection of PD disease-related modules co-expressed genes. (A) Outliers were detected in the sample cluster. (B) The cut-off point was set as 0.9, and the soft threshold power was set as β=8. (C) Tree diagram of all DEGs based on the cluster of difference measurement. The colored bands show the results from the automated monolithic analysis. (D) Correlation diagram between modules obtained by clustering according to inter-gene expression levels. (E) Heat map of the correlation between module characteristic genes and phenotypes. We chose the MEblack module for subsequent analysis (the ordinate value is the correlation coefficient of feature module). (F) Module membership vs. gene significance, cor is disease correlation coefficient, P is the P value of correlation coefficient. ME, module eigengene; WGCNA, weighted gene co-expression network analysis; PD, Parkinson’s disease; DEGs, differentially expressed genes.