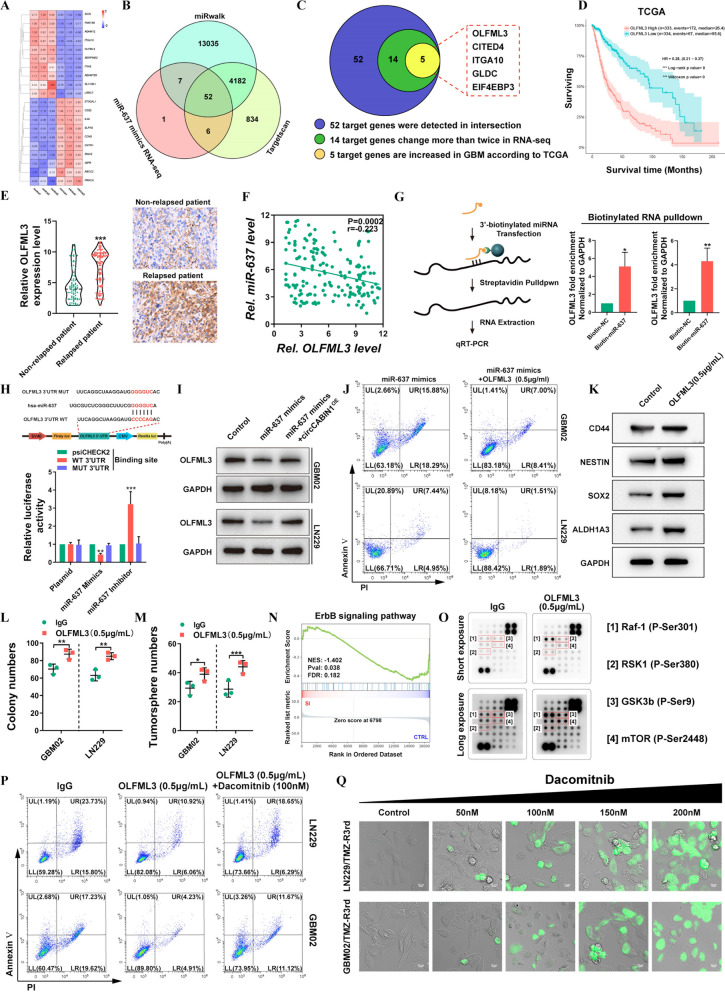
Fig. 6.
OLFML3 was a downstream target of the circCABIN1/miR-637 signalling axis. A-C 5 possible target genes were screened through RNA-seq, miRwalk, Targetscan and TCGA database. Heat map was performed to analyze the changed genes after transfected with miR-637 mimics (A). The venn diagram showed that 52 genes were changed together in three groups (B). Schematic illustration of genes significantly increased in GBM (C). D Kaplan–Meier analysis of OS in the high and low OLFML3 groups in TCGA. (p < 0.001). E The expression of OLFML3 in relapsed or non-relapsed patient and representative images of IHC in GBM sections of indicated groups. F Pearson correlation analysis of OLFML3 and miR-637 (p = 0.0002). G Pull down and qRT-PCR assay verified miR-637 binding with OLFML3 mRNA. H Schematic diagram of the OLFML3 mRNA and miR-637 binding sites and luciferase reporter assay verified OLFML3 mRNA binding with miR-637. I The expression of OLFML3 after transfected with miR-637 mimics alone or miR-637 mimics plus circCABIN1OE were analyzed by western blot in LN229 cells. J GBM cells administrated with miR-637 mimics alone or miR-637 mimics plus OLFML3 recombinant protein (0.5 μg/mL) were subjected to FACS to detected apoptosis. K-M Western blot, colony formation and tumorsphere formation assays detected the stem cell properties after treated with OLFML3 recombinant protein (0.5 μg/mL). N GSEA analysis showed that differential genes were mainly enriched in ErbB signal pathway (p = 0.038). O Protein array assay detected ErbB-related signalings after OLFML3 recombinant protein treatment (0.5 μg/mL). P GBM cells administrated with OLFML3 recombinant protein (0.5 μg/mL) alone or OLFML3 recombinant protein plus Dacomitnib (100 nM) were subjected to FACS to detected apoptosis. Q TMZ-resistance cells treated with dacomitnib with concentration gradient, and apoptosis was detected by caspase3/7 experiment. Scale bar, 10 μm. Results are presented as mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001