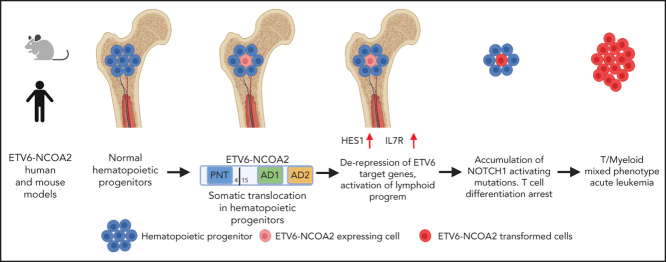
Abstract
Mixed-phenotype acute leukemia is a rare subtype of leukemia in which both myeloid and lymphoid markers are co-expressed on the same malignant cells. The pathogenesis is largely unknown, and the treatment is challenging. We previously reported the specific association of the recurrent t(8;12)(q13;p13) chromosomal translocation that creates the ETV6-NCOA2 fusion with T/myeloid leukemias. Here we report that ETV6-NCOA2 initiates T/myeloid leukemia in preclinical models; ectopic expression of ETV6-NCOA2 in mouse bone marrow hematopoietic progenitors induced T/myeloid lymphoma accompanied by spontaneous Notch1-activating mutations. Similarly, cotransduction of human cord blood CD34+ progenitors with ETV6-NCOA2 and a nontransforming NOTCH1 mutant induced T/myeloid leukemia in immunodeficient mice; the immunophenotype and gene expression pattern were similar to those of patient-derived ETV6-NCOA2 leukemias. Mechanistically, we show that ETV6-NCOA2 forms a transcriptional complex with ETV6 and the histone acetyltransferase p300, leading to derepression of ETV6 target genes. The expression of ETV6-NCOA2 in human and mouse nonthymic hematopoietic progenitor cells induces transcriptional dysregulation, which activates a lymphoid program while failing to repress the expression of myeloid genes such as CSF1 and MEF2C. The ETV6-NCOA2 induced arrest at an early immature T-cell developmental stage. The additional acquisition of activating NOTCH1 mutations transforms the early immature ETV6-NCOA2 cells into T/myeloid leukemias. Here, we describe the first preclinical model to depict the initiation of T/myeloid leukemia by a specific somatic genetic aberration.
Key Points
-
•
ETV6-NCOA2 expression in human and mouse hematopoietic nonthymic progenitors induces T/myeloid leukemia.
-
•
ETV6-NCOA2 induces a lymphoid program in hematopoietic progenitors through de-repression of ETV6 targets.
Introduction
T/myeloid mixed-phenotype acute leukemia (MPAL) is defined by the expression of myeloid and T-lymphoid markers on the same leukemic blast. MPAL is characterized by poor prognosis,1 and clinical management is challenging.2 Genomic studies revealed mutually exclusive alterations in the transcriptional regulators WT1, ETV6, RUNX1, and CEBPA in 82% of patients with T/myeloid MPAL2 in addition to alterations in transcriptional regulators.2, 3, 4, 5, 6 Despite the accumulating evidence from genomic analysis, the pathogenesis of T/myeloid MPAL is largely unknown because of the lack of preclinical models.
T-cell development is a continuous process in which hematopoietic stem cells differentiate into mature T cells.7 Activation of Notch1 in early prethymic progenitors (ETPs) ensures lymphoid priming8 and represses B-cell and myeloid genes in the thymus through sequential stages that are defined by the presence of specific cell surface molecules (ETP/DN1-DN4).9 Consequently, the cells lose their pluripotency and differentiate into mature T cells.10, 11 The mutational landscape of T/myeloid MPAL and its genomic similarity to hematopoietic stem cells and early progenitor cells suggest that the leukemic initiating event occurs in early hematopoietic precursors rather than in T-cell precursors.2 Yet, this has never been functionally demonstrated in preclinical models.
ETV6-NCOA2 is a fusion gene created by the t(8;12)(q13;p13) chromosomal translocation associated with T/myeloid leukemia,12, 13 which is often accompanied by somatic NOTCH1-activating mutations. The ETV6-NCOA2 fusion protein consists of the ETV6 N-terminal pointed (PNT) homo- and heterodimerization domain and the NCOA2 C-terminal activation domains AD1 and AD2, which interact with acetyltransferases and arginine methyltransferases, respectively (Figure 1A). Both ETV6 and NCOA2 are involved in chromosomal translocations in hematopoietic malignancies. Dominant negative ETV6 mutations were reported in immature adult T-cell acute lymphoblastic leukemia (T-ALL),14 whereas the AD1 and AD2 domains of NCOA2 are retained in a chromosomal translocation with lysine acetyltransferase 6A (KAT6A-NCOA2, MOZ-TIF2) associated with acute myeloid leukemia (AML).15, 16, 17 The recruitment of histone acetyltransferases by the HAT domain of KAT6 and the AD1 domain of NCOA2 is essential for myeloid transformation by KAT6-NCOA2.18 Because ETV6-NCOA2 and KAT6A-NCOA2 differ only in their N-terminal partner, it is reasonable to hypothesize that these partners are responsible for the specific leukemia immunophenotypes.
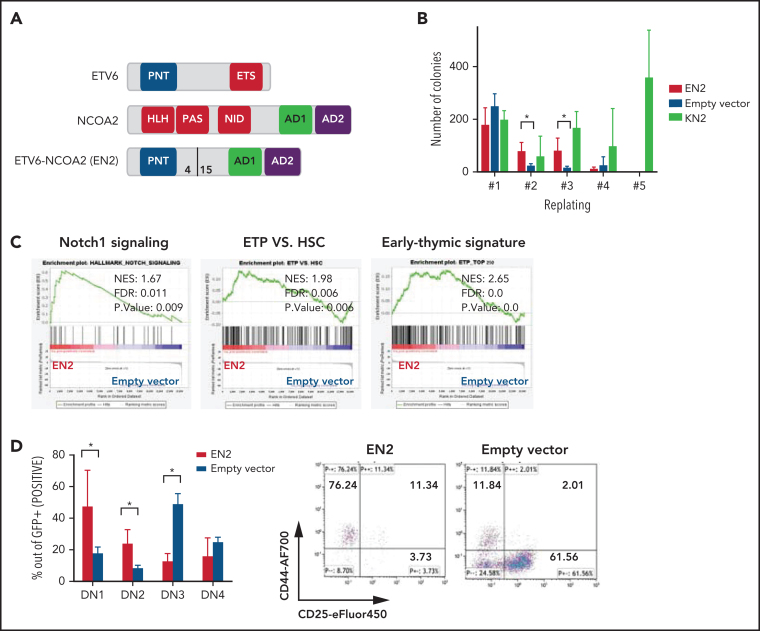
Figure 1.
ETV6-NCOA2 is a T-cell oncogene that induces immature T-cell arrest in murine BM progenitors. (A) Schematic representation of ETV6-NCOA2 (EN2) fusion. bHLH-PAS, basic helix-loop-helix Per-ARNT-SIM domain; NID, nuclear receptor interaction domain. (B) Murine BM progenitors treated with fluorouracil were transduced with MSCV-MIGR1-IRES-GFP retroviruses expressing ETV6-NCOA2, empty vector, or KAT6A-NCOA2 (KN2) and plated in methylcellulose (IL-3, IL-6, and SCF). Colonies were counted and re-plated every 7 to 10 days. EN2 induces self-renewal of transduced cells on methylcellulose culture compared with the empty vector–transduced cells in the second and third replate (Mann-Whitney U test P = .014) (n = 3). (C) Lineage-negative cells (lin–) were enriched from wt-C57BL/6 mice; the cells were transduced with EN2, KN2, or empty vector, incubated in liquid culture (IL-3, IL-6, and SCF) for 5 days, and sorted for GFP+. The RNA of the GFP+ sorted cells was sent for bulk RNA-seq. Gene set enrichment analysis (GSEA) of the EN2 vs empty vector–transduced cells demonstrated enrichment of Notch1 signature (NES, 1.67; FDR, 0.011), ETP (NES, 1.98; FDR, 0.006), and early thymic signature (NES, 2.65; FDR, 0.0). (D) Murine BM HSPCs treated with fluorouracil were transduced with either EN2 or empty vector and plated on OP9-DL4 stroma (IL-7, Flt3L) for 3 weeks and then immunophenotyped by flow cytometry (n = 4). Left panel: average immunophenotype results (Mann-Whitney U test for EN2 vs empty vector P = .02 in DN1, DN2, and DN3). Right panel: representative example of CD44 and CD25 flow cytometry results. AF700, Alexa Fluor 700; HSC, hematopoietic stem cell; SS, side scatter.
The tight association between a somatic genetic aberration and a T/myeloid leukemia phenotype provides an opportunity to study the pathogenesis of this leukemia. Here we used ETV6-NCOA2 to generate the first mouse and human T/myeloid leukemia model. Our results indicate that ETV6-NCOA2 expression induces de-repression of ETV6 target genes in nonthymic hematopoietic progenitors and drives the cells into the T-cell developmental trajectory, while withholding their full differentiation through preservation of the myeloid program.
Materials and methods
Vectors
ETV6-NCOA2 expression vectors MigR1-IRES-GFP and MigR1-IRES-NGFR contain a Flag tag. ETV6 expression vector MigR1-IRES-GFP contains a hemagglutinin (HA) tag. NOTCH1-L1601PdP-IRES-GFP expression vectors were kindly provided by Warren S. Pear (University of Pennsylvania, Philadelphia, PA). KAT6A-NCOA2-IRES-GFP was kindly provided by Brian Huntly (University of Cambridge, Cambridge, United Kingdom).
Enrichment of mouse and human progenitor cells
Bone marrow (BM) was harvested from the femurs and tibias of 6- to 8-week-old C57BL/6 mice. Mouse BM progenitor cells were enriched by treatment with fluorouracil 150 μg/g 48 hours before harvesting or by selection using the magnetic Lineage Cell Depletion Kit (Miltenyi Biotech). Enriched cells were cultured in RPMI supplemented with mouse interleukin-3 (mIL-3) 10 ng/mL, mIL-6 10 ng/mL, mouse stem cell factor (mSCF) 50 ng/mL (PeproTech), and cortisol 0.1 µM (Sigma). For human progenitor cells, anonymized cord blood units were obtained from Sheba Medical Center Cord Blood Bank under protocols approved by the Institutional Review Board. CD34+ cells were isolated using magnetic beads (Miltenyi Biotech) and cultured in Iscove modified Dulbecco medium supplemented with human stem cell factor, human thrombopoietin, and human FLT3L 100 ng/mL (PeproTech). For each biological replicate, at least 3 different cord blood units were mixed. Retroviral transduction (infection) of primary hematopoietic progenitors was performed using RetroNectin and double spinfection (centrifuged at 1800 rpm at 32°C for 60 minutes). To create patient-derived xenografts (PDXs), human BM samples were obtained according to the Declaration of Helsinki. The cells were engrafted in NOD-SCID IL2rγnull (NSG) mice.
Assays
For the methylcellulose re-plating assay, 3000 transduced cells per construct were plated in duplicate in methylcellulose supplemented with mIL-3, hIL-6, and mSCF (STEMCELL Technologies) in 24-well plates. Cells were incubated at 37°C in 5% CO2 for 7 to 10 days. Colonies with more than 50 cells were counted, and 3000 cells were replated into fresh methylcellulose cultures. For the OP9-DL4 T-cell differentiation assay, OP9-DL4 stromal cells (kindly provided by Ana Cumano, Pasteur Institute, Paris, France) were maintained at 37°C in 5% CO2 in opti-MEM ([Minimal Essential Medium]; Gibco), and 10 000 cells per well were pre-seeded in 12-well dishes. Then, 3000 transduced cells per well were plated in 1 mL opti-MEM supplemented with 1 ng/μL mIL-7 and 5 ng/μL mFlt3L (PeproTech). Every 3 days, each well was filtered using a 0.45-µm strainer, and cells were re-plated on fresh OP9-DL4 stroma. After 3 weeks, the cells were analyzed by flow cytometry for green fluorescent protein (GFP), CD4, CD8, CD25, CD44, Thy1.2, and cKit. For the transduction-transplantation assays, human or mouse hematopoietic progenitors were selected as described above. The transduction efficiency of each construct was evaluated by flow cytometer. For the luciferase reporter assay, 293T cells were retrovirally infected with the MSCV-pGL2-T574-IRES-GFP plasmid,14 and positive cells were selected using puromycin. Stably transduced cells were seeded (104 cells per well) in 24-well plates and transfected with 200 ng wild-type (wt) ETV6, ETV6-NCOA2 (200, 300, or 400 ng) (jetPEI; Polyplus) or their combination and were treated with A485 1 μM, A485 5 μM, A486 1 μM or A486 5 μM (kindly provided by Structural Genomics Consortium, Toronto, ON, Canada). Luciferase activity was measured at 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega).
Mouse strains
Six- to 8-week-old recipient C57BL/6 female mice (Envigo, Jerusalem, Israel) received lethal 6.5 Gy X-ray irradiation 24 hours before transplantation. Then, 2 × 105 transduced cells and 2 × 105 whole BM supporting cells (freshly harvested from 6-week-old C57BL/6 mice) were injected into the tail vein of irradiated recipients. Six- to 8-week-old NSGS (NOD.Cg-Prkdcscid Il2rgtm1WjlTg (CMV-IL3, CSF2, KITLG) female mice (The Jackson Laboratories) received sublethal 0.5 to 1 Gy X-ray irradiation 24 hours before transplantation. A total of 1 × 105 transduced cells or patient-derived cells were injected into the tail vein of irradiated recipients.
Flow cytometry and detection of Notch1 mutation
Cells were stained with fluorochrome-conjugated antibodies using standard protocols and were analyzed by using a Gallios flow cytometer (Beckman-Coulter). Data were analyzed using Kaluza software (Beckman-Coulter) (supplemental Methods, available on the Blood Web site). Genomic DNA was purified from the BM or spleens of ETV6-NCOA2 mice using a DNeasy Blood & Tissue Kit (QIAGEN). To amplify the HD and PEST domains, polymerase chain reactions were performed as previously described.12
RNA purification, cDNA preparation, and sequencing
Total RNA was extracted and purified using the TRIzol Plus RNA Purification Kit (Invitrogen). Complementary DNA (cDNA) was prepared by using the Verso cDNA Kit (Thermo Fisher Scientific). Bulk RNA sequencing (RNA-seq) was performed by first sorting the cells (5 × 105 cells per sample) using a BD FACSAria flow sorter. Library preparation and sequencing were performed by either the Nancy and Stephen Grand Israel National Center for Personalized Medicine research center of the Weizmann Institute of Science or the University of Cincinnati's sequencing core (supplemental Methods). H3K27Ac chromatin immunoprecipitation sequencing (ChIP-seq) was performed in: (1) human CD45+ cells from the BM of ETV6-NCOA2 PDX mice were sorted or (2) CD34+ cells that were enriched, transduced with ETV6-NCOA2 or empty vector, and cultured in vitro for 5 days. Sample preparation for ChIP-seq of H3K27Ac was performed as described before19 (supplemental Methods).
Co-IP
Coimmunopreipitation (Co-IP) was performed on a nuclear extract of DND41 cells stably expressing ETV6-NCOA2-Flag or ETV6-Flag (1 × 106 cells per sample) and on 293T cells transfected with ETV6-NCOA2-Flag or ETV6-HA. The lysate was added to 50 μL Dynabeads Protein-G (Thermo Fisher Scientific) bound to an antibody. The samples were analyzed by western blot (supplemental Methods).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8.1.2. Comparisons between 2 groups were performed by using an unpaired Student t test. The Mann-Whitney U test and the one-way nonparametric analysis of variance (ANOVA) were used when Gaussian distribution could not be assumed. The log-rank (Mantel-Cox) test was used for survival curves. Values of P < .05 were considered statistically significant.
Results
ETV6-NCOA2 is a T-cell oncogene that induces immature T-cell arrest in murine BM progenitors
The ETV6-NCOA2 fusion gene created by the t(8;12)(q13;p13) chromosomal translocation was first identified in T/myeloid MPAL in pediatric patients; the fusion was the sole karyotype abnormality in several patients.12 Therefore, we hypothesized that the ETV6-NCOA2 fusion protein is a transforming oncogene. To test this hypothesis, we first assessed the ability of ETV6-NCOA2 to self-renew. Murine BM hematopoietic stem and progenitor cells (HSPCs) were retrovirally infected with ETV6-NCOA2 (EN2), empty vector, or KAT6A-NCOA2 (KN2, MOZ-TIF2) as a positive control for NCOA2 fusion that induces AML.16 Self-renewal was tested by using the serial re-plating assay. Starting from the second replating, ETV6-NCOA2–transduced cells demonstrated higher clonogenic potential than did empty vector–transduced cells. After the third replating, ETV6-NCOA2–transduced cells were exhausted, whereas KAT6A-NCOA2–transduced cells continued to re-plate (Figure 1B). This result indicates that ETV6-NCOA2–expressing HSPCs have a mild self-renewal advantage.
To identify the primary changes in gene expression induced by ETV6-NCOA2 or KAT6A-NCOA2 in murine BM HSPCs,20, 21 we performed bulk RNA-seq after 5 days of in vitro culture (IL-3, IL-6, and SCF). We identified 238 genes with significant differential expression in ETV6-NCOA2–transduced HSPCs (56 upregulated and 182 downregulated genes) and 798 genes with differential expression in KAT6A-NCOA2–transduced HSPCs (146 upregulated and 652 downregulated genes). GSEA revealed enrichment of the Notch1 pathway, upregulation of Hes1, and induction of a T-cell signature in ETV6-NCOA2–transduced progenitors but not in KAT6A-NCOA2 (Figure 1C; supplemental Figures 1 and 2).
Because ETV6-NCOA2 expression in patients has been associated with the early T-cell phenotype,12 we hypothesized that ETV6-NCOA2 expression disrupts T-cell differentiation. To examine this, ETV6-NCOA2 or empty vector–transduced murine BM HSPCs were plated on OP9-DL4 stroma and followed for T-cell differentiation. ETV6-NCOA2–transduced cells were arrested at the early stages of T-cell differentiation CD4–CD8– DN1 (CD44+CD25–) or DN2(CD44+CD25+), whereas the empty vector–transduced cells differentiated to DN3(CD44–CD25+) (Figure 1D). ETV6-NCOA2 cells had a lower percentage of Thy1.2 expression and a higher percentage of cKit expression than the empty vector (supplemental Figure 3). Thus, ETV6-NCOA2 expression in murine BM HSPCs induces a T-cell program but arrests T-cell differentiation at an early stage; this is consistent with the T/myeloid phenotype of ETV6-NCOA2 leukemias.
ETV6-NCOA2 induces a T/myeloid hematopoietic malignancy accompanied by spontaneous Notch1 mutations in C57BL/6 mice
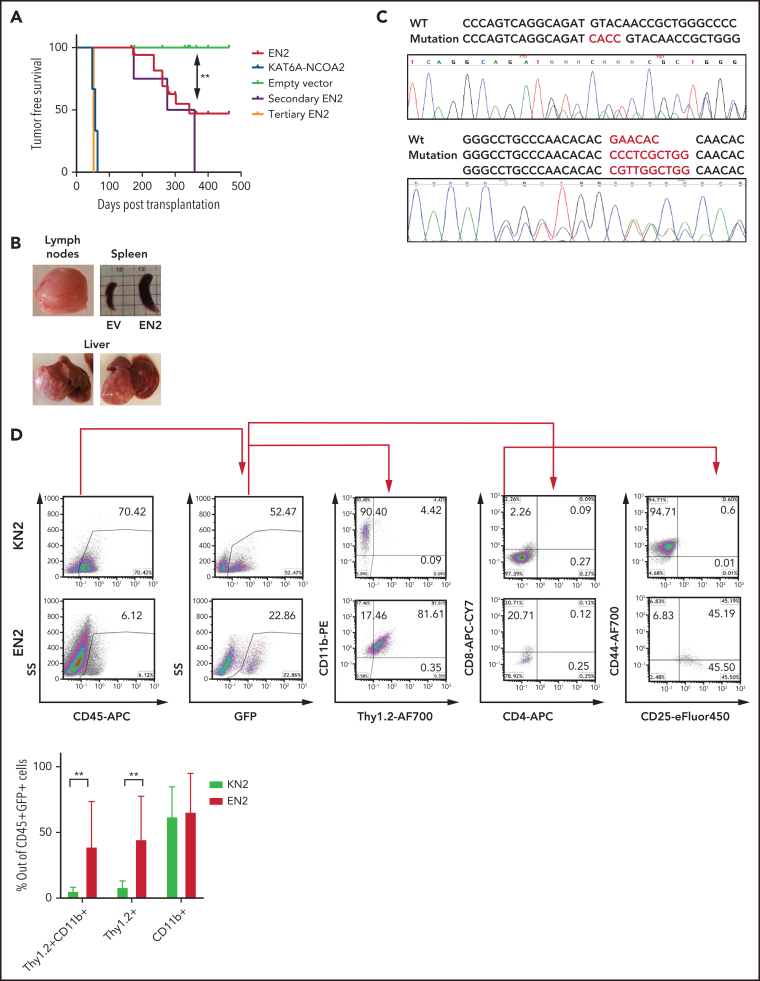
To assess the in vivo transforming properties of ETV6-NCOA2, murine BM HSPCs transduced with ETV6-NCOA2 (n = 18), KAT6A-NCOA2 (n = 3), or empty vector (n = 16) were transplanted into C57BL/6 mice. As expected, mice transplanted with KAT6A-NCOA2 developed AML after a median of 57 days,16, 18 and the mice that were transplanted with empty vector–transduced HSPCs did not develop disease. Half the mice transplanted with ETV6-NCOA2 developed hematopoietic malignancies; the median time to tumor development was 340 days (log-rank test ETV6-NCOA2 vs empty vector P = .0016) (Figure 2A). The mice developed lymphomas with nodular infiltration of the liver, spleen, and lymph nodes. Hematoxylin and eosin staining demonstrated extramedullary hematopoiesis, atypical and abnormal lymphomatous infiltration of the hematopoietic organs, and destruction of their normal architecture (Figure 2B; supplemental Figure 4; supplemental Table 1). Even though most patients with this translocation develop leukemia, 1 patient with ETV6-NCOA2 T-lymphoblastic lymphoma with myeloid antigens was reported.22 Secondary ETV6-NCOA2 transplanted mice (n = 4) developed hematopoietic malignancies at a median of 317 days after transplantation. Tertiary transplanted mice (n = 2) developed T or T/M leukemia with high BM infiltration after 52 days (Figure 2A; supplemental Figure 5; supplemental Table 2). Consistent with the T-cell gene induction observed in the in vitro assays of ETV6-NCOA2–transduced HSPCs, ETV6-NCOA2–driven tumors displayed an early T-cell phenotype with myeloid markers. The ETV6-NCOA2 population was enriched with DN1 to DN3 cells with CD11b expression (Figure 2C). This immature immunophenotype is similar to the phenotype of ETV6-NCOA2 human leukemia.12
Figure 2.
ETV6-NCOA2 induces a T/myeloid hematopoietic malignancy accompanied by spontaneous Notch1 mutations in C57BL/6 mice. C57BL/6 mice BM progenitors treated with fluorouracil were transduced with MSCV-MIGR1-IRES-GFP retroviruses expressing EN2 (n = 18), KN2 (n = 3), or empty vector (EV) (n = 16). (A) Kaplan-Meier tumor-free survival analysis of EN2 primary, secondary (n = 4), and tertiary (n = 2) mice compared with KN2 and empty vector (log-rank test EN2 vs empty vector P = .0016). (B) Examples of EN2 tumors. (C) Flow cytometry analysis of KN2- and EN2-transduced leukemic cells. Top: flow examples of CD45, GFP, CD11b/Thy1.2, CD4/CD8, and CD44/CD25. Bottom: summary of Thy1.2 and CD11b flow cytometry results (Mann-Whitney U test **P = .0095). (D) Two examples of Notch1 mutations in the PEST domain detected in EN2 leukemia/lymphoma in C57BL/6 mice. APC, allophycocyanin; PE, phycoerythrin.
The course of ETV6-NCOA2 disease in C57BL/6 mice was characterized by partial penetrance and long latency, a trend that meets the need for accumulation of additional somatic mutations. Because mutations in NOTCH1 HD and PEST domains are common in patients with ETV6-NCOA2,12, 23 tumors from mice transplanted with ETV6-NCOA2 were screened for similar mutations. Indeed, spontaneous mutations in the PEST domain were found in 40% of the mice that were transplanted with ETV6-NCOA2–transduced HSPCs (Figure 2D; supplemental Table 1); these mice presented with higher intracellular Notch1 compared with nonmutated mice (supplemental Figure 6). These transplantation results demonstrate that ETV6-NCOA2 is a driving event in the murine hematopoietic system, causing T/myeloid hematopoietic malignancies.
ETV6-NCOA2 collaborates with NOTCH1 to induce T/myeloid leukemia in human HSPCs
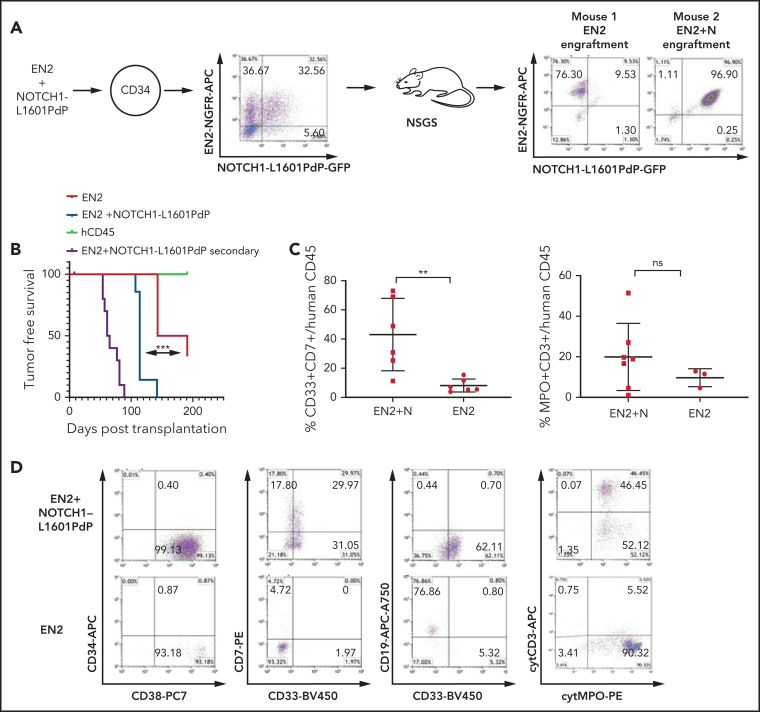
To explore the effect of ETV6-NCOA2 expression on the initiation of T/myeloid MPAL in human hematopoietic cells, CD34+ cord blood cells were transduced with ETV6-NCOA2. Because NOTCH1 was found to be a progression event in ETV6-NCOA2–driven malignancies, as demonstrated in human patients and in the C57BL/6 mouse model, CD34+ cord blood progenitor were co-transduced with an activating mutant NOTCH1-L1601PdP and ETV6-NCOA2. NOTCH1-L1601PdP has a mutation in the HD domain and deletion of the PEST domain, similar to the NOTCH1 mutations identified in human T-ALL.24 The cotransduced ETV6-NCOA2-NGFR and NOTCH1-L1601PdP-GFP cells were transplanted into NSGS mice25 (n = 15). This coinfection generated 4 distinct populations that competed within the mice and constituted intrinsic controls (Figure 3A): (1) ETV6-NCOA2 + NOTCH1-L1601PdP, (2) ETV6-NCOA2, (3) NOTCH1-L1601PdP, and (4) untransduced CD34+ cells. Postmortem analysis of the BM cells revealed that 7 of the 15 transplanted mice engrafted with the cotransduced ETV6-NCOA2 + NOTCH1-L1601PdP cells had a median survival of 114 days. Six of the 15 mice engrafted with ETV6-NCOA2–transduced cells had a median survival of 167 days (log-rank ETV6-NCOA2 vs ETV6-NCOA2 + NOTCH1-L1601PdP P = .0004). Two healthy mice were engrafted with untransduced human CD45 cells. NOTCH1-L1601PdP–transduced cells did not engraft in the NSGS mice, further emphasizing the requirement for cooperativity between the 2 oncoproteins (supplemental Figure 7). BM cells from ETV6-NCOA2 + NOTCH1-L1601PdP engrafted mice (n = 4) or ETV6-NCOA2 engrafted mice (n = 4) were transplanted into 3 to 4 NSGS recipients from each donor mouse. The mice transplanted with ETV6-NCOA2 + NOTCH1-L1601PdP double-positive cells developed leukemia in secondary engrafted mice at 50 to 90 days after transplantation (Figure 3B).
Figure 3.
ETV6-NCOA2 collaborates with NOTCH1 to induce T/myeloid leukemia in human CD34+ progenitors. Human cord blood CD34+ progenitors were co-transduced with MSCV-MIGR1-IRES retroviruses that express EN2-NGFR and NOTCH1-L1601PdP-GFP. (A) The cotransduction generated 4 groups of human cells that were cotransplanted into NSGS recipient mice (n = 15): (1) EN2, (2) NOTCH1-L1601PdP, (3) EN2 + NOTCH1-L1601PdP, and (4) untransduced CD34+ cells; right: examples of 2 mice, one with EN2 engraftment and the other with EN2 + NOTCH1-L1601PdP Log-rank (Mantel-Cox) (N) engraftment. (B) Kaplan-Meier tumor-free survival for primary and secondary mice by postmortem engraftment analysis (EN2 vs EN2 + NOTCH1-L1601PdP; P = .0004). (C) Summary of EN2 and EN2 + NOTCH1-L1601PdP immunophenotype CD33/CD7 (left) and cytMPO/cytCD3 (right) (Mann-Whitney U test; **P = .0087). Panel C represents the percentage of positive cells out of the CD45+ population. Each dot represents a mouse. (D) Examples of flow cytometry analysis of EN2-NGFR and EN2 + NOTCH1-L1601PdP-GFP, CD34/CD38, CD7/CD33, CD19/CD33, and cytCD3/cytMPO. Cyt, cytoplasmic; ns, not significant.
All ETV6-NCOA2 + NOTCH1-L1601PdP double-transduced cells displayed a T/myeloid mixed immunophenotype with either CD34–CD38+CD7+cytCD3+CD33–/MPO+ (5 of 7 mice) or CD34+CD38+CD7+cytCD3+CD33+/MPO– (2 of 7 mice). This variation in myeloperoxidase (MPO) expression resembles the partial MPO expression in human patients with ETV6-NCOA2.12 Mice engrafted with single-positive ETV6-NCOA2–transduced cells displayed B-cell and myeloid markers (Figure 3D; supplemental Table 3). Thus, ETV6-NCOA2 collaborates with activation of NOTCH1 to transform human nonthymic CD34+ progenitors into early immature T/myeloid leukemia.
ETV6-NCOA2 induces a transcriptional program in human cord blood hematopoietic progenitors, which is similar to primary patient-derived ETV6-NCOA2 leukemias
ETV6-NCOA2 protein is a fusion of 2 transcriptional regulators, so we hypothesized that ETV6-NCOA2 transforms hematopoietic progenitors by altering gene expression. We examined the global transcriptomic profile of ETV6-NCOA2–induced cells at different stages of disease development: human CD34+ cells transduced with ETV6-NCOA2 or empty vector in vitro, ETV6-NCOA2 and ETV6-NCOA2 + NOTCH1-L1601PdP–engrafted cells in NSGS mice, and PDXs generated from ETV6-NCOA2 pediatric leukemias (supplemental Figure 8).
Analysis of ETV6-NCOA2 CD34+ in vitro transduced cells demonstrated enrichment with early lymphoid genes,26 early thymic progenitors,27 and ETP signatures14 (supplemental Figure 9A-C), similar to the gene signature induced by ETV6-NCOA2 transduction in murine hematopoietic progenitors (Figure 1C). NOTCH1 target HES1 was significantly upregulated, as was the myeloid receptor CSF1R, which was found to have a role in lymphoid-myeloid checkpoint.28 In addition, significant activation of KRAS signaling was detected at this early stage, as previously described for ETP-ALL2 (supplemental Figure 9D).
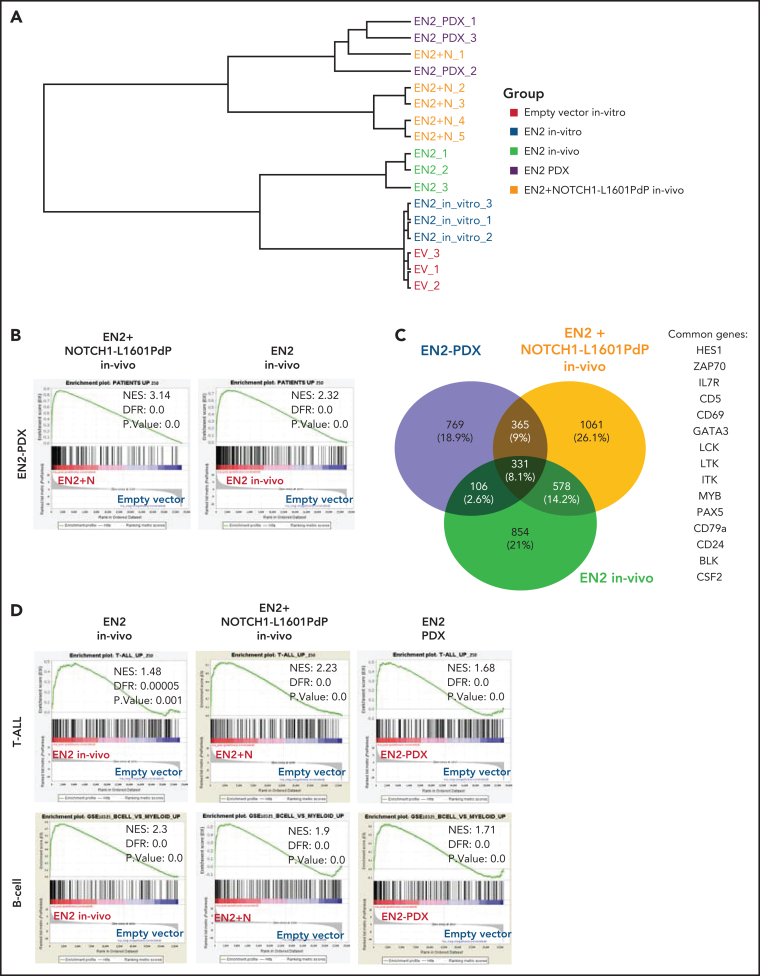
We have shown that co-expression of ETV6-NCOA2 + NOTCH1-L1601PdP induces T/myeloid leukemia. To explore the mechanism and the effect of NOTCH1-L1601PdP on ETV6-NCOA2–induced gene expression, we compared the in vivo ETV6-NCOA2–engrafted cells and ETV6-NCOA2 + NOTCH1-L1601PdP leukemia with ETV6-NCOA2 PDXs. Unsupervised hierarchical clustering ordered the samples into 2 branches according to the presence of NOTCH1 mutations. One branch comprised expression of ETV6-NCOA2 in vitro and in vivo, whereas the other comprised ETV6-NCOA2 + NOTCH1-L1601PdP in vivo samples and the ETV6-NCOA2 PDX samples, which stresss the relevance of the model to human leukemia (Figure 4A).
Figure 4.
ETV6-NCOA2 is a transcriptional regulator that induces a gene expression pattern similar to that in ETV6-NCOA2 pediatric leukemia. Human cord blood CD34+ progenitors were transduced with MSCV-MIGR1-IRES-GFP retroviruses expressing EN2 or empty vector. The cells were cultured in vitro (SCF, FLT3L, and thrombopoietin (TPO) for 5 days and sorted for GFP+. In vivo EN2 engrafted cells (n = 3) were sorted for CD45+NGFR+, EN2 + NOTCH1-L1601PdP leukemia cells were sorted for CD45+GFP+NGFR+ (n = 3), and EN2 PDX cells were sorted for CD45+ (n = 3) from BM of transplanted sick NSG mice. The sorted cells were sent for bulk RNA-seq. (A) Unsupervised clustering of the samples based on the 500 top differentially expressed genes in the RNA-seq experiments. (B) GSEA analysis of EN2 + NOTCH1-L1601PdP (left) and EN2 (right) compared with the 250 top-ranked upregulated genes of human EN2 PDXs. The differentially expressed genes of the EN2 and EN2 + NOTCH1-L1601PdP samples were ranked according to their log10 (P value) and compared with the patient's gene set. (C) Venn diagram of significantly upregulated genes in the human in vivo EN2 engrafted cells, in vivo EN2 + NOTCH1-L1601PdP leukemia, and EN2 PDX samples. (D) GSEA pre-ranked analysis of in vivo EN2–engrafted cells, in vivo EN2 + NOTCH1-L1601PdP leukemia, and EN2 PDX samples compared with the T-ALL gene set (top) or B-cell gene set (bottom).
We generated an ETV6-NCOA2 upregulated gene signature list by comparing ETV6-NCOA2 PDXs to CD34+ cells transduced with empty vector ranking in the 250 top significantly upregulated genes. GSEA of both ETV6-NCOA2 + NOTCH1-L1601PdP leukemia (normalized enrichment score [NES], 3.14; false discovery rate [FDR], 0) and ETV6-NCOA2–engrafted cells (NES, 2.32; FDR, 0.0) showed significant positive enrichment of the ETV6-NCOA2 PDX signature. The correlation score for the ETV6-NCOA2 PDX gene set was higher for ETV6-NCOA2 + NOTCH1-L1601PdP than for ETV6-NCOA2, which supports the significance of NOTCH1 in ETV6-NCOA2 leukemogenesis (Figure 4B).
The common group of significantly upregulated genes in the in vivo ETV6-NCOA2–engrafted cells, ETV6-NCOA2 + NOTCH1-L1601PdP leukemia, and ETV6-NCOA2 PDXs includes 331 genes that are enriched with lymphoid genes (IL7RA, GATA3, HES1, LCK, PAX5, CD79A, and BLK) (Figure 4C), which define ETV6-NCOA2 as a lymphoid driver (Figure 4D). Consistent with the immunophenotyping of the cells (Figure 3D), ETV6-NCOA2–engrafted cells express B-cell genes (CD19 and IGLV/C) and were negative for NOTCH1 mutations, whereas ETV6-NCOA2 + NOTCH1-L1601PdP leukemia and ETV6-NCOA2 PDXs expressed T-cell genes (eg, CD7).
MEF2C is highly expressed in early immature T-ALL13, 29, 30 as well as in normal human thymocyte subsets; however, in normal thymocytes, its expression is significantly decreased beyond the DN2 stage.13, 27, 28, 29, 30, 31 MEF2C is expressed in ETV6-NCOA2 PDX cells13 (supplemental Figure 9). The T-cell differentiation and the T/myeloid checkpoint are dependent on accurate regulation of NOTCH1 and PU.1/MEF2C pathways.32, 33, 34, 35 For an early thymocyte to differentiate into a mature T cell, the NOTCH1 pathway should be activated whereas the PU.1/MEF2C pathway should be completely shut down (PU.1, MEF2C, and CSF1R).32, 33, 34, 35, 36 Together these results indicate that ETV6-NCOA2 is a lymphoid transcriptional activator that collaborates with NOTCH1 to drive extrathymic hematopoietic progenitors into the T-cell trajectory. This occurs while maintaining expression of myeloid genes, thus arresting the cells at the early immature T-cell differentiation stage.
ETV6-NCOA2 causes de-repression of ETV6 targets
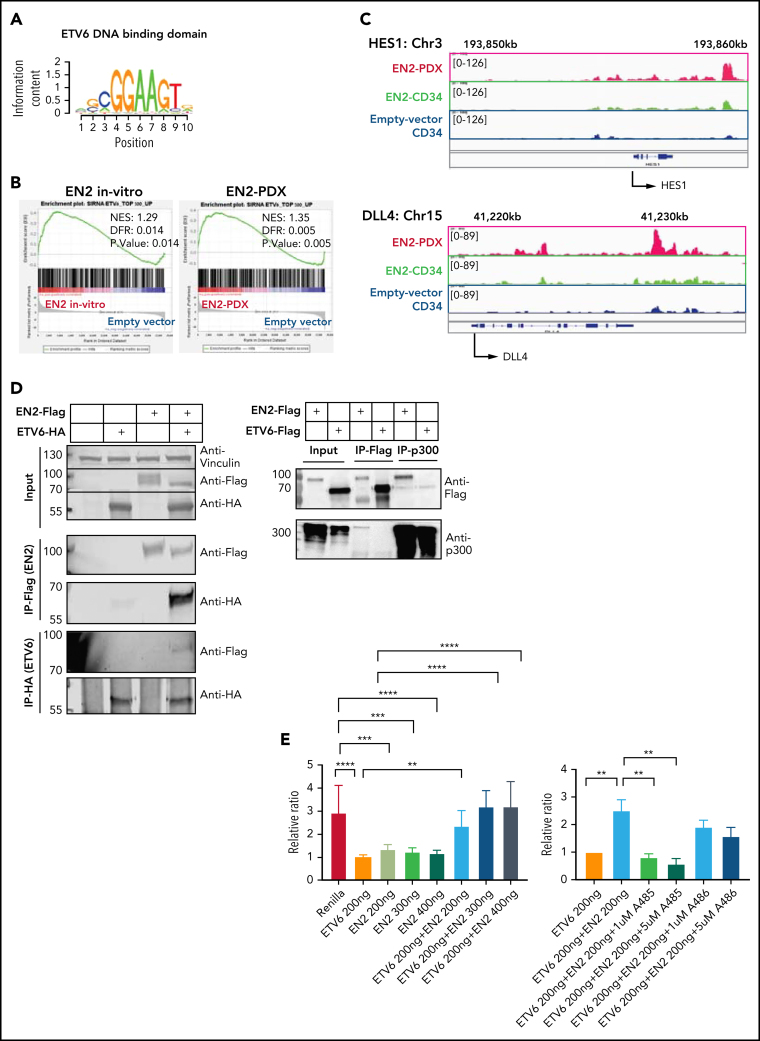
DNA-binding site analysis37 of the significant upregulated genes in the RNA-seq demonstrated enrichment of the erythroblast transformation–specific (ETS) 5GCGGAAGT3 binding motif in all of the ETV6-NCOA2 samples: ETV6-NCOA2 CD34+ in vitro transduced cells (NES, 4.77), in vivo ETV6-NCOA2–engrafted cells (NES, 3.56), ETV6-NCOA2+NOTCH1-L1601PdP leukemia (NES, 4.43), and ETV6-NCOA2 PDXs (NES, 3.46) (Figure 5A; supplemental Table 5).37, 38, 39 However, this enrichment was not observed in the downregulated genes.
Figure 5.
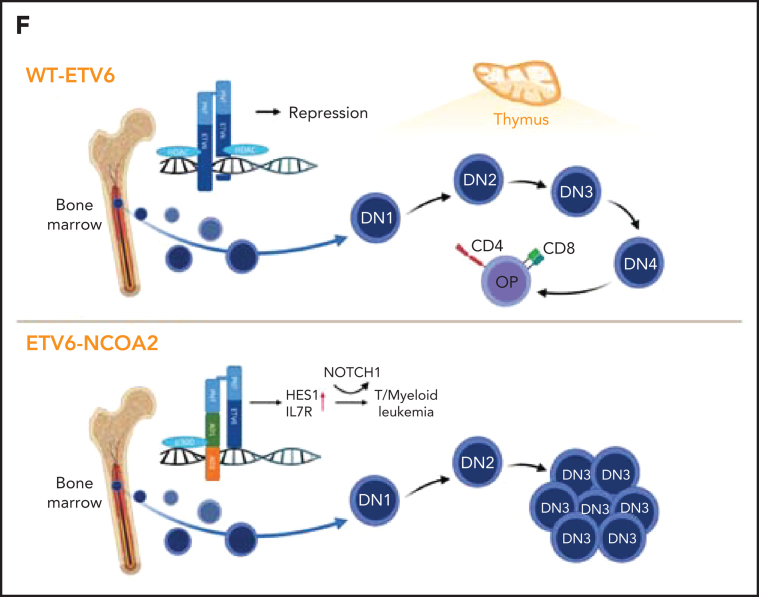
ETV6-NCOA2 causes de-repression of ETV6 targets. (A) Transcription factor binding site analysis (i-Cis target37). The analysis was performed separately for the significantly upregulated and downregulated differentially expressed genes of the bulk RNA-seq of CD34+ cells transduced with EN2 or empty vector, and for the in vivo EN2 engrafted cells, in vivo EN2 + NOTCH1-L1601PdP leukemia, and EN2 PDX samples. (B) GSEA analysis of EN2 in vitro and EN2 PDX sample pre-ranked RNA-seq results compared with the 500 top upregulated genes in the LOUCY cell line treated with short interfering RNA (siRNA) against ETV6 (siETV6).14 (C) ChIP-seq for H3K27Ac was performed on CD34+ cells transduced with EN2 or empty vector in vitro, and EN2 PDX. H3K27 differential acetylation of HES1 and DLL4 was examined by integrative genomics viewer63 analysis. (D) Left: Co-IP EN2/ETV6: HEK-293T cells were transfected with empty vector, EN2-Flag, ETV6-HA, or a combination of both. The proteins were immunoprecipitated with either Flag antibody (EN2) or HA antibody (ETV6). Right: Co-IP EN2/p300: EN2-Flag or ETV6-Flag stable DND41 cells were immunoprecipitated with either Flag antibody (top) or endogenous p300 antibody (bottom). (E) Left: 293T cells stably expressing ETV6 luciferase reporter vector were transfected with 200 ng ETV6 alone, increasing concentrations of EN2 (200, 300, and 400 ng) or a combination of both (ANOVA test P < .0001) (n = 6). Right: ETV6-luciferase reporter stable transfected 293T cells were transfected with 200 ng ETV6 alone or with 200 ng ETV6 with 1 of the following: EN2 200 ng, EN2 + 1 200 ng, or 5 μM A485 (a p300 inhibitor)51 or EN2 EN2 200 ng in addition to 1 or 5 μM A485 or A486 (inactive A485 analog) (ANOVA test P < .0001). (F) A scheme for proposed EN2 mechanism of leukemogenesis. Upper panel: ETV6-HDAC repression of genes in wt hematopoietic progenitors. Lower panel: EN2-ETV6-p300 complex in EN2 hematopoietic progenitors. This figure was created with BioRender.com. **P < .008; ***P < .0006; ****P < .0001. Chr, chromosome; HDAC, histone deacetylase.
Because ETV6 is a transcriptional repressor,40 the above observations could suggest that ETV6-NCOA2 causes de-repression of ETV6 target genes. We confirmed this by comparing the ETV6-NCOA2 expression profile to a published gene signature of siETV6 in the early T-cell leukemia LOUCY cell line.14 These comparisons demonstrated a significant positive correlation in all of the following groups: ETV6-NCOA2 CD34+ in vitro transduced cells (NES, 1.29; FDR, 0), in vivo ETV6-NCOA2–engrafted cells (NES, 1.65; FDR, 0.0), ETV6-NCOA2+NOTCH1-L1601PdP leukemia (NES, 1.58; FDR, 0), and ETV6-NCOA2 PDXs (NES, 1.35; FDR, 0.005)14 (Figure 5B; supplemental Figure 10A).
To validate that ETV6-NCOA2 reverses the repression of ETV6 target genes, we combined the gene expression of ETV6-NCOA2 CD34+ in vitro transduced cells with a published ENCODE ChIP-seq of ETV6 in K562 cells,41 a hematopoietic myelogenous leukemia cell line,42 by binding and expression target analysis (BETA).43 Significant enrichment of the ETS family of transcription factors was observed in the peaks of the up-target (Student t test P = 1.7e−16; T score, 8.41) (supplemental Figure 10b). ETV6 de-repressed genes included HES1 and DLL4, which are activators in the NOTCH1 pathway.44 Both were significantly upregulated in RNA-seq and have H3K27 acetylation peaks in ETV6-NCOA2 PDXs (Figure 5C). This suggests that ETV6 de-repression is involved in early T-cell programming initiated by ETV6-NCOA2 in nonthymic hematopoietic progenitors.
Loss of the wt-ETV6 allele is a prevalent event in leukemias with ETV6 translocation (eg, ETV6-RUNX1).45, 46, 47, 48 However, ETV6-NCOA2 cells preserve their wt-ETV6, as we previously demonstrated.12 ETV6-NCOA2 lacks the DNA-binding domain of ETV6 but maintains the PNT homo- and heterodimerization domain.49 Dominant negative mutations in ETV6 with an intact PNT dimerization domain were described in ETP-ALL.14 Hence, we hypothesized that ETV6-NCOA2 interacts with wt-ETV6. To verify this interaction, we performed co-IP experiments. ETV6-NCOA2-Flag and ETV6-HA were pulled down with Flag or HA antibody, respectively. We observed that ETV6-NCOA2 and ETV6 are pulled down together, thus creating a protein complex (Figure 5D).
NCOA2 is a transcriptional coactivator that recruits p300 H3 acetyltransferase through its AD1 domain, which is retained in the ETV6-NCOA2 and KAT6A-NCOA250 fusions. This interaction was proposed to have an important role in KAT6A-NCOA2–induced leukemogenesis.18, 50 To explore the interaction between ETV6-NCOA2 and p300, we expressed ETV6-NCOA2-Flag or ETV6-Flag in DND41 cells and pulled down either Flag-tagged protein or endogenous p300.18 Co-IP of ETV6-NCOA2-Flag pulled down p300, whereas the immunoprecipitation of ETV6-Flag did not. Together, these experiments suggest the formation of an ETV6-NCOA2–wt-ETV6-p300 complex (Figure 5D).
To validate ETV6 derepression by ETV6-NCOA2, we performed a luciferase reporter assay. We generated a stably expressing pGL2-T574 luciferase reporter 293T cell line with an ETV6 DNA recognition site.13 wt-ETV6 repressed the basal transcription level of this promoter. Expression of ETV6-NCOA2 in the cells together with wt-ETV6 activated the promoter in a dose-dependent manner (Figure 5E) (ANOVA P < .0001). A485, a p300 inhibitor, reduced the H3K27Ac levels of both ETV6-NCOA2 and empty vector cells (supplemental Figure 11). Treating the wt-ETV6 cells with A485 derepressed the transcription from the promoter, but its inactive analog A486 did not51 (ANOVA P < .0001) (Figure 5E). Interestingly, the expression of ETV6-NCOA2 alone caused repression of the transcription from the reporter gene. Similar to KAT6A-NCOA2, ETV6-NCOA2 (which lacks a DNA-binding domain) binds p300 and CREB-binding protein, thus reducing its accessibility in the cell. Additional evidence for that is the general H3K27Ac reduction in ETV6-NCOA2 cells compared with that in empty vector (supplemental Figure 12).
On the basis of these results, we propose that ETV6-NCOA2 is recruited to ETV6 target genes by heterodimerization with wt-ETV6. The recruitment of p300 histone acetyltransferase to NCOA2 AD1 activates ETV6-repressed lymphoid genes in nonthymic hematopoietic progenitors. The acquisition of activating NOTCH1 mutations and preservation of the expression of myeloid genes results in T/myeloid leukemias (Figure 5F).
Discussion
T/myeloid leukemias, clinically classified as either MPAL or ETP, are characterized by coexpression of T-cell and myeloid cell markers and poor prognosis.2, 52 Consistent with the T/myeloid immunophenotype, genomic analysis revealed somatic mutations typical to both T-ALL and AML.2 Yet the pathogenesis of T/myeloid ALL is largely unknown, and preclinical models are lacking. On the basis of the previous observations12, 13 that ETV6-NCOA2 is tightly associated with T/myeloid leukemias, we hypothesized that the ETV6-NCOA2 fusion gene is an oncogene that induces T/myeloid leukemia.
Two preclinical models confirmed this hypothesis; ectopic expression of ETV6-NCOA2 in mouse HSPCs led to T/myeloid lymphomas associated with acquisition of spontaneous Notch1–activating mutations. Similarly, coexpression of ETV6-NCOA2 and non-transforming activated NOTCH1 mutant (NOTCH1-L1601PdP)24 in human cord blood CD34+ cells resulted in aggressive T/myeloid leukemia in immunodeficient mice. Remarkably, we showed that the expression of ETV6-NCOA2 in both mouse and human HSPCs, even though cultured for a short time under myeloid conditions in vitro, induced gene expression of a lymphoid program and the arrest of T-cell differentiation in DN1 and DN2 on OP9-DL4 stroma. Our study suggests that the initiation of the lymphoid program in extrathymic HSPCs, coupled with preservation of myeloid gene expression and acquisition of NOTCH1 mutations, leads to T/myeloid leukemias.
NCOA2 is involved in additional recurrent translocation in leukemias; KAT6-NCOA2 (MOZ-TIF2) causes aggressive AML.53 Mechanistic studies suggested a dual leukemogenic mechanism. The first is depletion of the histone acetyltransferase p300 by its binding to the NCOA2 AD1 domain.15, 18, 54 The second complementary mechanism is the specific recruitment of p300 to promoters of myeloid genes such as CSF1 thereby increasing their expression.55
We propose a similar mechanism for the initiation of T/myeloid leukemia by ETV6-NCOA2 in extrathymic hematopoietic progenitors. In ETV6-NCOA2, the same segment of NCOA2 is fused to the PNT domain of ETV6. The known role of the ETV6 PNT domain is to mediate ETV6 homodimerization.49 Indeed, we showed that ETV6-NCOA2 forms a complex with ETV6, thereby recruiting p300 to ETV6 targets and reversing their repression, inducing a mostly lymphoid program, as we have observed in the reporter (Figure 5E) and gene expression experiments (Figure 4C-D). Furthermore, a general reduction in H3K27Ac levels was detected in ETV6-NCOA2–expressing cells, suggesting a depletion of p300 by ETV6-NCOA2. Interestingly, p300 is often inactivated in ETP and T/myeloid leukemias2, 56
The T-cell differentiation checkpoint depends on the downregulation of myeloid genes, specifically MEF2C and PU.1. NCOA2 is a known regulator of MEF2C.57 Indeed, we observed expression of MEF2C upon expression of ETV6-NCOA2. Homminga et al13 demonstrated that MEF2C is a key transcription factor in the lymphoid-myeloid lineage decision; its expression is high in B cells and T-lymphoid progenitors; however, it dramatically decreased beyond the DN2 stage16, 25, 26, 27 during T-cell development. Consistent with our findings, MEF2C was upregulated in an immature T-ALL cohort that included 3 patients with ETV6-NCOA2.29, 30
We found that ETV6-NCOA2 induces B and T genes in extrathymic hematopoietic progenitors, yet it leads to T/myeloid leukemia. It is possible that the lack of RAG1/2 expression in these progenitors prevented the deletions of B-cell differentiation genes that are almost universally observed in B-cell precursor ALL.58 Interestingly, expression of B-cell antigens has been described in T-ALL,59 ETV6-NCOA2 leukemias13, 30 (Jules P. P. Meijerink, Prinses Máxima Center for Pediatric Oncology, Utrecht, The Netherlands. The communication was via email, 6 May 2021), and MEF2C-expressing ETPs.29, 60 Expression of a competing lineage gene is common in B-cell and T-cell leukemias, possible affecting their differentiation arrest (eg, the T-cell gene GATA3 in B-cell precursor ALL61, 62).
Although ETV6-NCOA2 is a rare somatic event in leukemia, the genetic mechanism described here is of general significance for the pathogenesis of T/myeloid MPAL. Our novel preclinical models suggest that T/myeloid leukemias are initiated in early hematopoietic progenitors. This is consistent with the recent genomic analysis of T/myeloid MPAL2 that reported a high frequency of somatic ETV6 mutations. Moreover, Van-Vlierberghe et al14 reported frequent dominant negative ETV6 mutations in T-ALL. Together, these publications support our observations that de-repression of ETV6 target genes in HSPCs is a general key initiating event of T/myeloid MPAL.
Acknowledgments
The authors thank Elizbeth Macintyre (Department of Immunology, Infectiology and Haematology, Institut Necker Enfants-Malades, Paris, France) and Keisuke Kato (Division of Pediatric Hematology and Oncology, Ibaraki Children's Hospital, Ibaraki, Japan) for providing samples from patients with ETV6-NCOA2; Warren S. Pear (University of Pennsylvania, Philadelphia, PA) for providing NOTCH1-IRES-GFP expression vectors; Brian Huntly (University of Cambridge, Cambridge, United Kingdom) for providing MOZ-TIF2-IRES-GFP; Anna Cumano (Pasteur Institute, Paris, France) for providing the OP9-DL4 cell line; the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, the Weizmann Institute of Science and Genomics, and the Epigenomics and Sequencing Core at the University of Cincinnati for sequencing service; Itzhak Ben Moshe for help with xenograft experiments; and all past and present members of S.I.'s research group for fruitful discussions and advice.
This work was supported by a grant from the Israel Science Foundation (ISF), by a Joint National Natural Science Foundation of China-ISF Research Grant, by grants from the Sheba-Cincinnati Children's Medical Center, Israeli Health Ministry, the Waxman Cancer Research Foundation, Hans Neufeld Stiftung, The Nevzlin Family Genomic Center Award, The Horowitz Foundation, the Dotan Center for Research of Hematological Malignancies, and The Israel Childhood Cancer Foundation (all to S.I.). This work was also supported by grants from the Funds for Scientific Research–Flanders (FWO; Odysseus Grant), Kinderkankerfonds (a non-profit childhood cancer foundation under Belgian law), and the European Research Council (StG-639784) (all to P.V.V.).
The computational resources and services used in this work were provided by the Flemish Supercomputer Center (funded by the FWO and the Flemish Government, Department of Economics, Science and Innovation.
G.F. is the David and Stacey Cynamon Research Fellow Chair in Genetics and Personalized Medicine. S.I. is the Dora and Giorgio Shapiro Chair of Hematological Malignancies.
This work was performed in partial fulfilment of the degree requirements for H.F., Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
Footnotes
The RNA-seq and ChIP-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and are accessible via GSE162338.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.F. and S.I. designed the study; H.F. performed most of the experiments; H.F., S.M., I.G., V.B., Y.K., I.G., E.K., A.R.-G., G.S., Y.B., B.K., J.C.M., and P.V.V performed experiments; H.F., S.N.-L., W.V.L., and G.F. analyzed sequencing data; S.S., J.S., A.F., and A.N. contributed reagents; and H.F. and S.I. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Supplementary Material
REFERENCES
- 1.Gerr H, Zimmermann M, Schrappe M, et al. Acute leukaemias of ambiguous lineage in children: characterization, prognosis and therapy recommendations. Br J Haematol. 2010;149(1):84–92. doi: 10.1111/j.1365-2141.2009.08058.x. [DOI] [PubMed] [Google Scholar]
- 2.Alexander TB, Gu Z, Iacobucci I, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562(7727):373–379. doi: 10.1038/s41586-018-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckstein OS, Wang L, Punia JN, et al. Mixed-phenotype acute leukemia (MPAL) exhibits frequent mutations in DNMT3A and activated signaling genes. Exp Hematol. 2016;44(8):740–744. doi: 10.1016/j.exphem.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Wang F, Morita K, et al. Integrative genomic analysis of adult mixed phenotype acute leukemia delineates lineage associated molecular subtypes. Nat Commun. 2018;9(1):2670. doi: 10.1038/s41467-018-04924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan L, Ping N, Zhu M, et al. Clinical, immunophenotypic, cytogenetic, and molecular genetic features in 117 adult patients with mixed-phenotype acute leukemia defined by WHO-2008 classification. Haematologica. 2012;97(11):1708–1712. doi: 10.3324/haematol.2012.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montefiori LE, Bendig S, Gu Z, et al. Enhancer hijacking drives oncogenic BCL11B expression in lineage ambiguous stem cell leukemia [published online ahead of print 8 June 2021]. Cancer Discov., doi:10.1158/2159-8290.CD-21-0145. [DOI] [PMC free article] [PubMed]
- 7.Zhou W, Yui MA, Williams BA, et al. Single-cell analysis reveals regulatory gene expression dynamics leading to lineage commitment in early T cell development. Cell Syst. 2019;9(4):321–337.e9. doi: 10.1016/j.cels.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen ELY, Thompson PK, Zúñiga-Pflücker JC. RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat Immunol. 2019;20(11):1456–1468. doi: 10.1038/s41590-019-0518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah DK, Zúñiga-Pflücker JC. An overview of the intrathymic intricacies of T cell development. J Immunol. 2014;192(9):4017–4023. doi: 10.4049/jimmunol.1302259. [DOI] [PubMed] [Google Scholar]
- 10.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13(6):427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 11.Labrecque N, Baldwin T, Lesage S. Molecular and genetic parameters defining T-cell clonal selection. Immunol Cell Biol. 2011;89(1):16–26. doi: 10.1038/icb.2010.119. [DOI] [PubMed] [Google Scholar]
- 12.Strehl S, Nebral K, König M, et al. ETV6-NCOA2: a novel fusion gene in acute leukemia associated with coexpression of T-lymphoid and myeloid markers and frequent NOTCH1 mutations. Clin Cancer Res. 2008;14(4):977–983. doi: 10.1158/1078-0432.CCR-07-4022. [DOI] [PubMed] [Google Scholar]
- 13.Homminga I, Pieters R, Langerak AW, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19(4):484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208(13):2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin H, Glass J, Blanchard KL. MOZ-TIF2 repression of nuclear receptor-mediated transcription requires multiple domains in MOZ and in the CID domain of TIF2. Mol Cancer. 2007;6(1):51. doi: 10.1186/1476-4598-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91(9):3127–3133. [PubMed] [Google Scholar]
- 17.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36(8):945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Deguchi K, Ayton PM, Carapeti M, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3(3):259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 19.van Galen P, Viny AD, Ram O, et al. A multiplexed system for quantitative comparisons of chromatin landscapes. Mol Cell. 2016;61(1):170–180. doi: 10.1016/j.molcel.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 22.Bond J, Touzart A, Nadal N, et al. Early thymic precursor-like lymphomatous presentation of the ETV6-NCOA2 translocation. Br J Haematol. 2018;181(3):392–394. doi: 10.1111/bjh.14579. [DOI] [PubMed] [Google Scholar]
- 23.Mansour MR, Linch DC, Foroni L, Goldstone AH, Gale RE. High incidence of Notch-1 mutations in adult patients with T-cell acute lymphoblastic leukemia. Leukemia. 2006;20(3):537–539. doi: 10.1038/sj.leu.2404101. [DOI] [PubMed] [Google Scholar]
- 24.Chiang MY, Xu L, Shestova O, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wunderlich M, Chou FS, Link KA, et al. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24(10):1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurenti E, Doulatov S, Zandi S, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14(7):756–763. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belyaev NN, Biró J, Athanasakis D, Fernandez-Reyes D, Potocnik AJ. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. Immunogenetics. 2012;64(8):591–604. doi: 10.1007/s00251-012-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seki M, Kimura S, Isobe T, et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017;49(8):1274–1281. doi: 10.1038/ng.3900. [DOI] [PubMed] [Google Scholar]
- 29.Colomer-Lahiguera S, Pisecker M, König M, et al. MEF2C-dysregulated pediatric T-cell acute lymphoblastic leukemia is associated with CDKN1B deletions and a poor response to glucocorticoid therapy. Leuk Lymphoma. 2017;58(12):2895–2904. doi: 10.1080/10428194.2017.1312383. [DOI] [PubMed] [Google Scholar]
- 30.Zuurbier L, Gutierrez A, Mullighan CG, et al. Immature MEF2C-dysregulated T-cell leukemia patients have an early T-cell precursor acute lymphoblastic leukemia gene signature and typically have non-rearranged T-cell receptors. Haematologica. 2014;99(1):94–102. doi: 10.3324/haematol.2013.090233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soulier J, Clappier E, Cayuela JM, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106(1):274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 32.Champhekar A, Damle SS, Freedman G, Carotta S, Nutt SL, Rothenberg EV. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev. 2015;29(8):832–848. doi: 10.1101/gad.259879.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgescu C, Longabaugh WJ, Scripture-Adams DD, et al. A gene regulatory network armature for T lymphocyte specification. Proc Natl Acad Sci U S A. 2008;105(51):20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothenberg EV, Scripture-Adams DD. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin Immunol. 2008;20(4):236–246. doi: 10.1016/j.smim.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140(6):1207–1219. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stehling-Sun S, Dade J, Nutt SL, DeKoter RP, Camargo FD. Regulation of lymphoid versus myeloid fate ‘choice' by the transcription factor Mef2c. Nat Immunol. 2009;10(3):289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- 37.Herrmann C, Van de Sande B, Potier D, Aerts S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 2012;40(15):e114. doi: 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei GH, Badis G, Berger MF, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29(13):2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imrichová H, Hulselmans G, Atak ZK, Potier D, Aerts S. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 2015;43(W1):W57–W64. doi: 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez RG, Carron C, Oury C, Gardellin P, Bernard O, Ghysdael J. TEL is a sequence-specific transcriptional repressor. J Biol Chem. 1999;274(42):30132–30138. doi: 10.1074/jbc.274.42.30132. [DOI] [PubMed] [Google Scholar]
- 41.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein E, Ben-Bassat H, Neumann H, et al. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer. 1976;18(4):421–431. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Sun H, Ma J, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8(12):2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB. Dynamic ligand discrimination in the Notch signaling pathway. Cell. 2018;172(4):869–880.e19. doi: 10.1016/j.cell.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hock H, Shimamura A. ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol. 2017;54(2):98–104. doi: 10.1053/j.seminhematol.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coniat MB, Poirel H, Leblanc T, Bernard OA, Berger R. Loss of the TEL/ETV6 gene by a second translocation in ALL patients with t(12;21) Leuk Res. 1999;23(10):895–899. doi: 10.1016/s0145-2126(99)00105-8. [DOI] [PubMed] [Google Scholar]
- 47.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15(3):162–174. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Barjesteh van Waalwijk van Doorn-Khosrovani S, Spensberger D, de Knegt Y, Tang M, Löwenberg B, Delwel R. Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene. 2005;24(25):4129–4137. doi: 10.1038/sj.onc.1208588. [DOI] [PubMed] [Google Scholar]
- 49.Kim CA, Phillips ML, Kim W, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20(15):4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17(2):507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lasko LM, Jakob CG, Edalji RP, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550(7674):128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huntly BJ, Shigematsu H, Deguchi K, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 54.Collins HM, Kindle KB, Matsuda S, et al. MOZ-TIF2 alters cofactor recruitment and histone modification at the RARbeta2 promoter: differential effects of MOZ fusion proteins on CBP- and MOZ-dependent activators. J Biol Chem. 2006;281(25):17124–17133. doi: 10.1074/jbc.M602633200. [DOI] [PubMed] [Google Scholar]
- 55.Aikawa Y, Katsumoto T, Zhang P, et al. PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat Med. 2010;16(5):580–585. doi: 10.1038/nm.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14(10):1209–1228. [PMC free article] [PubMed] [Google Scholar]
- 58.Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35(9):975–983. doi: 10.1200/JCO.2016.70.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mi X, Griffin G, Lee W, et al. Genomic and clinical characterization of B/T mixed phenotype acute leukemia reveals recurrent features and T-ALL like mutations. Am J Hematol. 2018;93(11):1358–1367. doi: 10.1002/ajh.25256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canté-Barrett K, van Helsdingen Y, Goossens S, Vroegindeweij E, Meijerink J. Conditional MEF2C expression in the mouse thymus drives development of T/B-biphenotypic lymphoid leukemia/lymphoma. HemaSphere. 2019;3:412. S1. [Google Scholar]
- 61.Walsh KM, de Smith AJ, Chokkalingam AP, et al. GATA3 risk alleles are associated with ancestral components in Hispanic children with ALL. Blood. 2013;122(19):3385–3387. doi: 10.1182/blood-2013-08-524124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Andreu V, Roberts KG, Harvey RC, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. 2013;45(12):1494–1498. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.