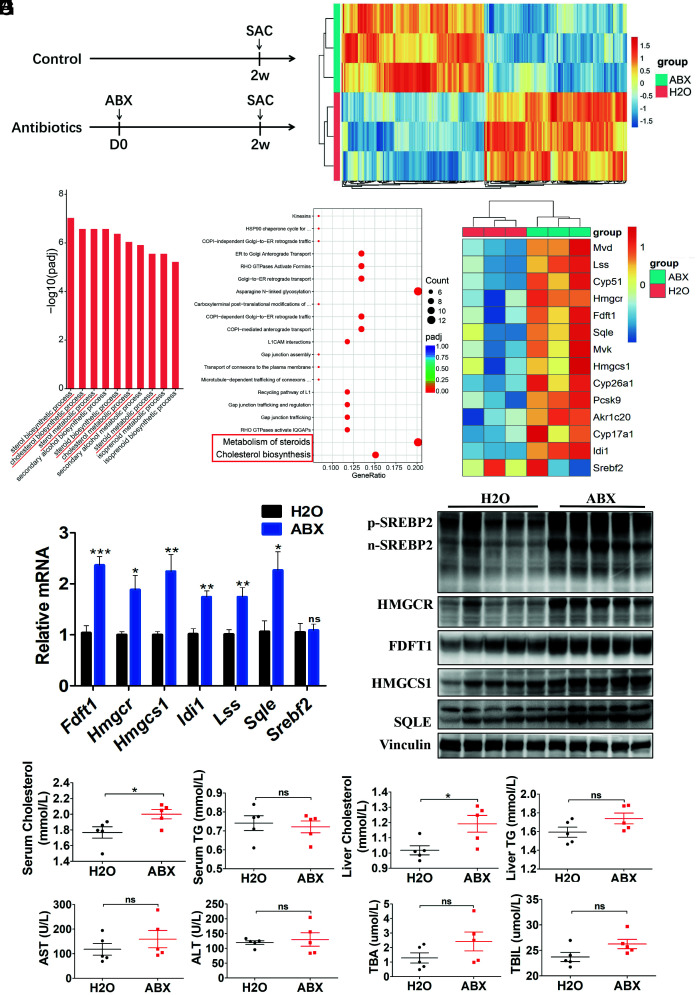
Fig. 3.
SREBP2 and downstream cholesterol synthesis-related gene expression increased in mice with gut commensal bacteria depletion. (A) Experimental procedure (n = 5 per group). Mice were treated with antibiotics or H2O for 2 wk and killed for further analysis. (B) Differentially expressed genes in normal liver tissues from mice in the ABX and H2O groups identified using transcriptome analysis (n = 3 per group). (C) Gene Ontology (GO) and (D) Reactome pathway enrichment analysis showing the cholesterol metabolism-related processes and pathways, respectively, indicated by red tags. (E) Transcriptome analysis of genes involved in cholesterol biosynthesis pathway. (F) Gene expression in liver tissues from ABX- or H2O-treated mice was determined by qRT-PCR (n = 5 per group). (G) Western blot analysis of liver tissues from ABX- or H2O-treated mice (n = 5 per group). (H) Serum and liver cholesterol and triglyceride levels in mice treated with ABX or H2O (n = 5 per group). (I) Serum AST, ALT, total bile acid (TBA), and total bilirubin (TBIL) levels in mice treated with ABX or H2O (n = 5 per group). Data were presented as mean ± SEM, P values were calculated by Student’s t test. ns, not significant; *P <0.05; **P <0.01; ***P <0.001.