Figure 5. H-H dimerization enables FoxP3 to recognize diverse sequences.
See also Figure S6.
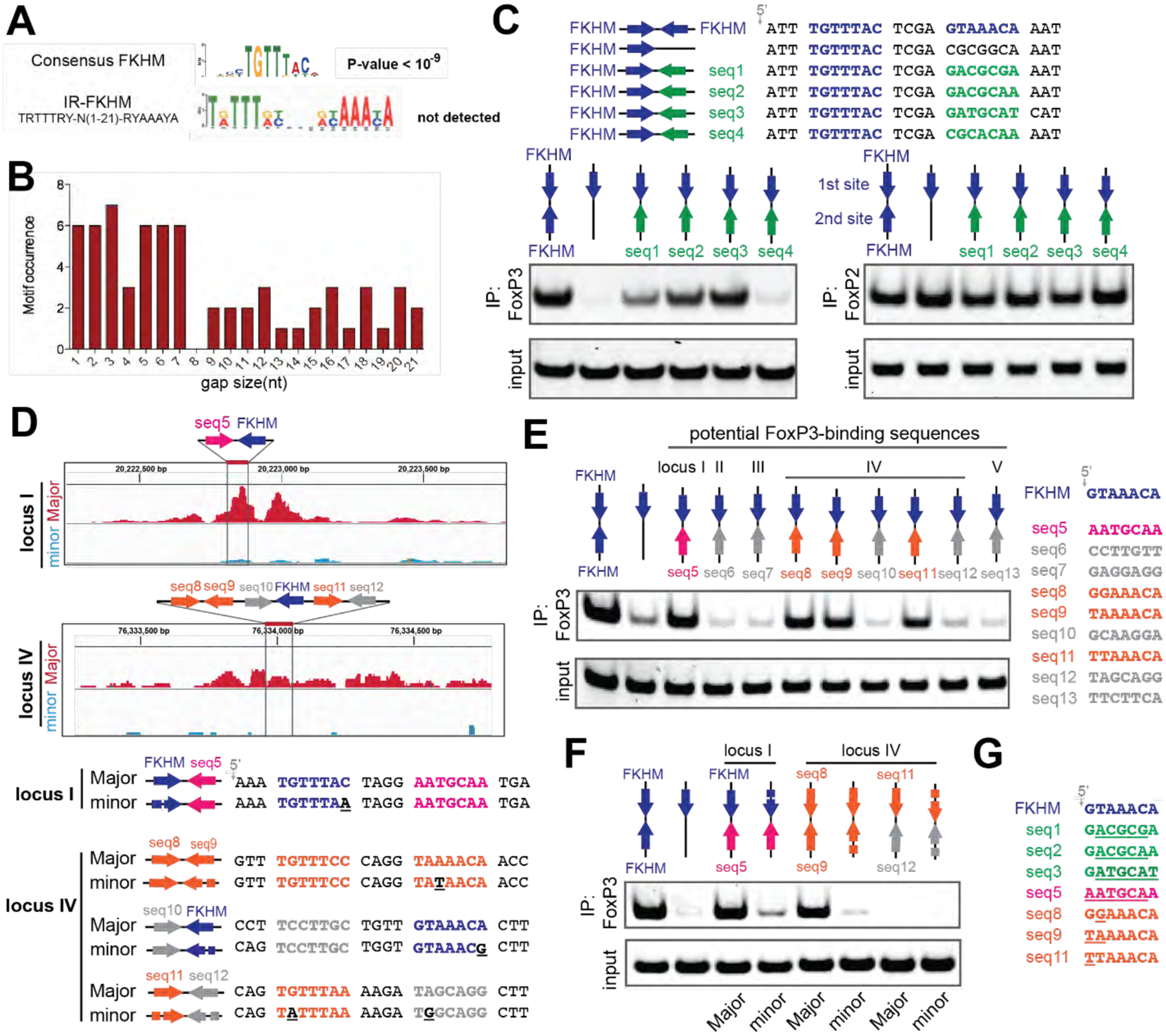
A. De novo motif analysis of FoxP3 ChIP-seq (n=5000) sequences. The canonical FKHD motif was enriched (P-value<10−9), but IR-FKHM motif was not detected for any of the gap sizes tested (1–21 nt). A more relaxed FKHM sequence (TRTTRY; R and Y indicate purine and pyrimidine, respectively) was used to be inclusive.
B. Occurrence of IR-FKHM, iteratively testing gap sizes of 1 to 21 nt, of FoxP3 ChIP-seq sequences that contain an FKHM (n=548). Motif occurrences were counted if p-value<10−5 and score greater than 12.
C. FoxP3 interaction with DNA harboring IR-FKHM4g and FKHM paired with non-canonical motifs (seq1–4). Purified MBP-FoxP3ΔN was mixed with DNA oligos and was subjected to MBP pull-down, followed by native PAGE analysis of co-purified DNA. DNA was visualized by Sybr Gold stain. Right: MBP-FoxP2ΔN was used for comparison.
D. FoxP3 Cut&Run intensity showing allelic imbalance of FoxP3 occupancy in loci I and IV. Major and minor alleles indicate alleles with greater and lesser FoxP3 occupancy, respectively. Below: Major and minor allele sequences with their differences highlighted with underscores in the minor sequences.
E. FoxP3 interaction with DNA harboring potential FoxP3-binding sequences paired with FKHM. These sequences were chosen from five loci in the Major allele sequence where mutations were associated with reduced FoxP3 occupancy.
F. FoxP3 interaction with DNA harboring natural sequences from loci I and IV (see D).
G. Alignment of FoxP3-compatible sequences examined in this figure. Sequence deviation from FKHM was highlighted with underscore.
Data in (C, E and F) are representative of at least three independent experiments.
