Figure 6. H-H dimerization is also important for Runx1 binding.
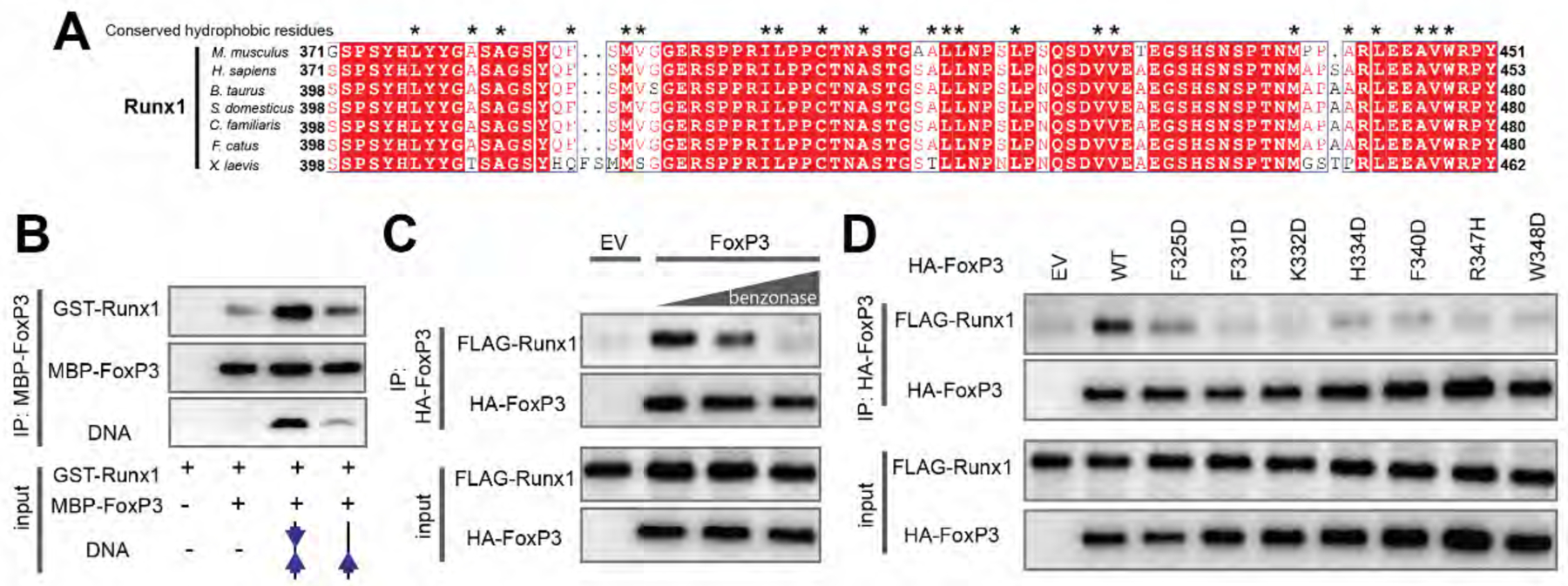
A. Sequence alignment of Runx1 orthologs showing conserved hydrophobic residues (*) in its C-terminal tail that binds FoxP3.
B. FoxP3 interaction with Runx1 using purified proteins. MBP-tagged FoxP3RBR-forkhead and GST-tagged Runx1 (residue 371–451) was purified from E. coli and was subjected to MBP pull-down in the presence and absence of DNA harboring IR-FKHM4g or single FKHM.
C. FoxP3 interaction with Runx1 from 293T cells. HA-tagged full-length FoxP3 and FLAG-tagged full-length Runx1 were separately expressed in 293T cells. Cell lysates were mixed and were subjected to anti-HA immunoprecipitation (IP). The increasing concentrations of benzonase was used to examine the effect of cellular DNA on FoxP3–Runx1 interaction.
D. FoxP3 interaction with Runx1 from 293T cells. H-H interface mutations were compared to WT FoxP3. Experiments were performed as in (C), except no benzonase was used in all conditions.
Data in (B-D) are representative of at least three independent experiments.
