Figure 6.
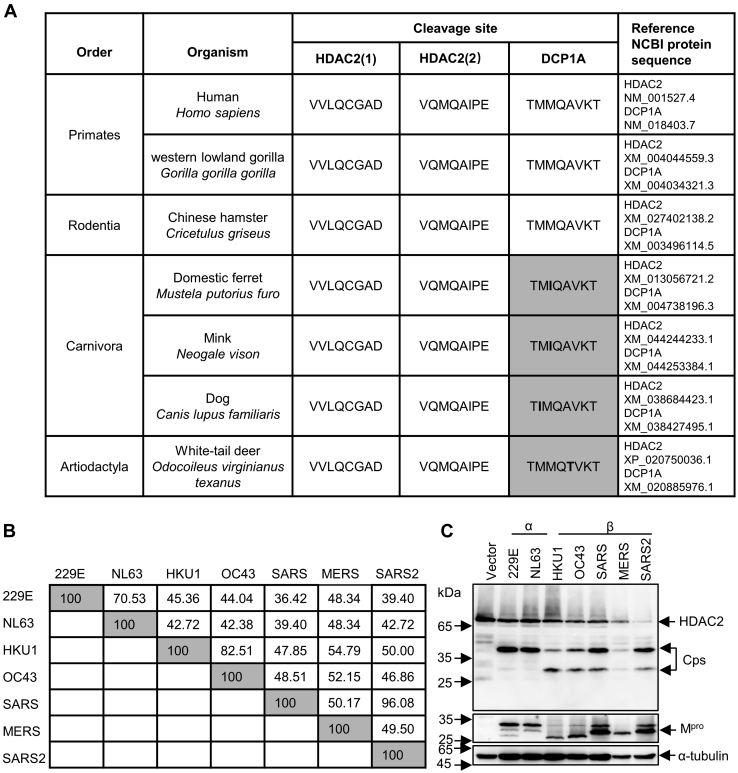
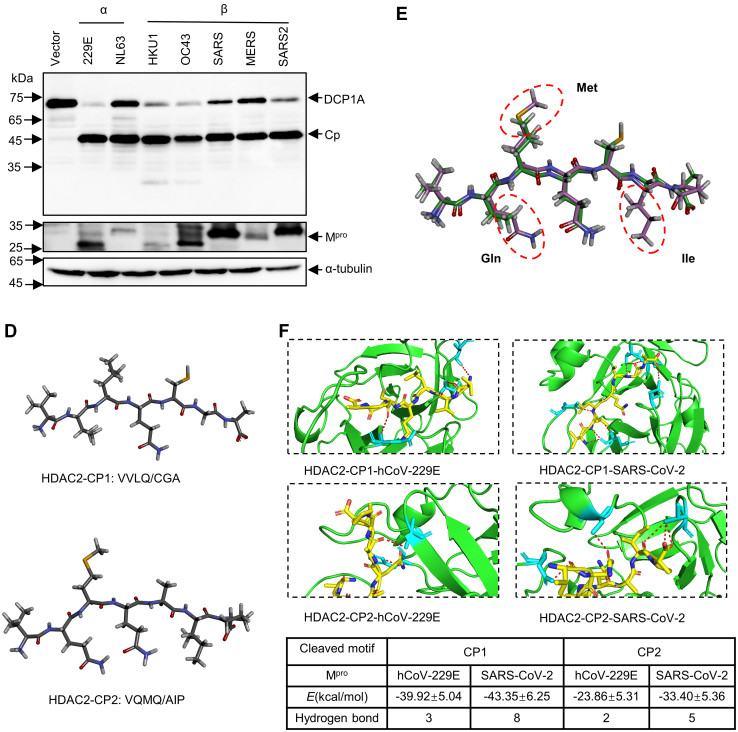
Mprocleavage efficiency differs in different coronaviruses.A, analysis of protein sequences of HDAC2 and DCP1A in cleaved sites across species. B, amino acid percent identity matrix of Mpro proteins in different coronaviruses. C, HEK-293T cells were individually transfected with empty vector, 229E-Mpro, NL63-Mpro, HKU1-Mpro, OC43-Mpro, SARS-Mpro, MERS-Mpro, or SARS2-Mpro plasmids along with HDAC2 or DCP1A. Western blotting was performed 30 h posttransfection. D, generation of peptide conformations based on sequence of two cleaved motifs in HDAC2. CP1 represents the first cleaved peptide, and CP2 means the second cleaved peptide. E, comparison of predicted two heptapeptides structures using superimpose in discovery studio. F, binding energy and hydrogen bonds formed between Mpro and cleaved motifs. The hydrogen bonds (Red) formed between the two cleaved peptides and Mpro are presented as dash lines labeled with donor residual (yellow) and corresponding acceptor residual (cyan) in Mpro. CP, cleaved peptide; DCP1A, mRNA-decapping enzyme 1a; HDAC, histone deacetylase; Mpro, main protease.