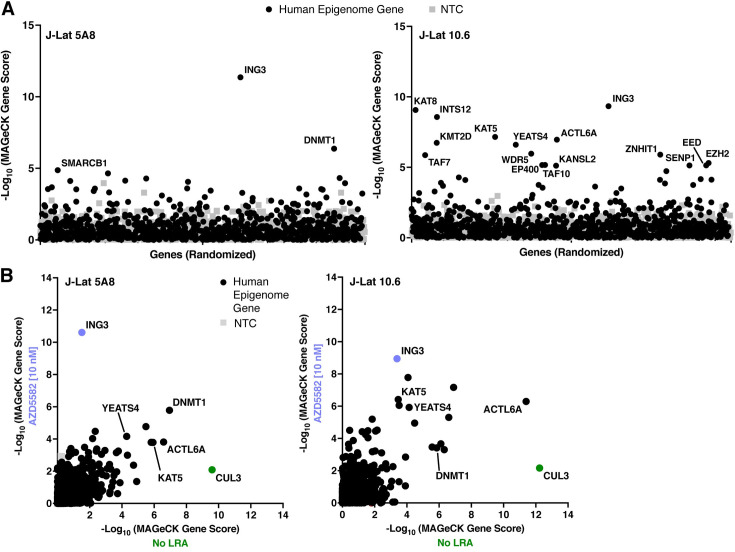
Fig 5. LRA Latency HIV-CRISPR screen identifies ING3 in combination with AZD5582 as a HIV-1 latency maintenance factor.
(A) ING3 is the top hit of the LRA Latency HIV-CRISPR screen. The Latency HIV-CRISPR screen HuEpi knockout cells were treated with a low activating dose (10 nM) of AZD5582. The -log10 of the MAGeCK score (on the y-axis) for each gene targeted by the HuEpi sgRNA library (black circles) and NTCs (gray squares) are calculated and displayed. Gene names are labelled for hits that have a <10% false discovery rate (FDR) in each J-Lat cell lines. NTCs are artificial NTC genes designed by iterative binning of NTC sgRNA sequences (see Methods). Genes are randomized on the x-axis, but the same order is used for both right and left panels. (B) Comparison of the Latency HIV-CRISPR screen by MAGeCK score in the presence (y-axis) and absence of AZD5582 (x-axis) with HuEpi genes (circles) and NTCs (gray squares). The data for the screen without an LRA (x-axis) is from Fig 3 as these two screens were performed in parallel. The genes unique to each screen are closest to the respective axis and the genes that are in common to both screens are at the center. ING3 is highlighted in periwinkle and CUL3 in green.