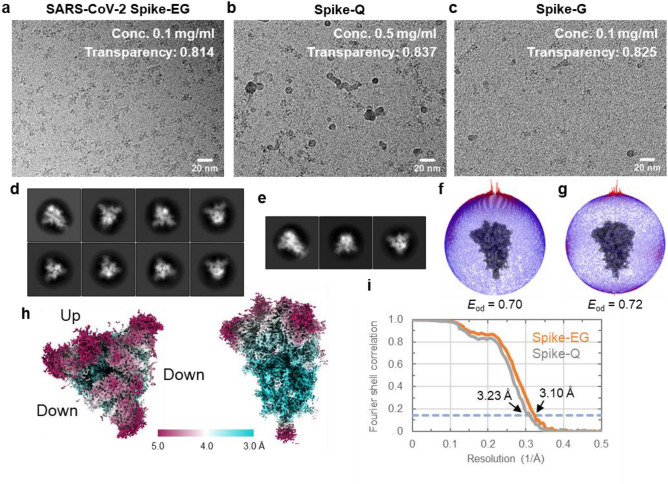
Figure 4.
CryoEM image analyses of SARS-CoV-2 spike protein on the EG-grid and Quantifoil grid. (a–c) Typical cryoEM images of spike protein on the EG-grid (spike-EG) (a), on the Quantifoil grid (spike-Q) (b), and on the glow discharged graphene grid (spike-G) (c). The protein concentration and the value of electron beam transparency of each image (measured from the image brightness ratio between the energy filter slit in and out) are also shown. (d and e) Selected 2D class averages from the spike-EG dataset (d) and the spike-Q dataset (e). The class averages are aligned in descending order of particle numbers from left to right and top to bottom. (f and g) Angular distributions of particles used in the final refinement for the spike-EG dataset (f) and the spike-Q dataset (g). Final 3D maps are shown for reference. The efficiencies (Eod) calculated with cryoEF are also shown. (h) Two orthogonal views of the final 3D map from the spike-EG dataset: top view of the trimer, left panel; and side view, right panel. The local resolution distribution is colored as in the color bar. (i) The FSC curves for the final maps of the spike-EG and spike-Q dataset. The dashed blue line indicates the FSC = 0.143 criterion.