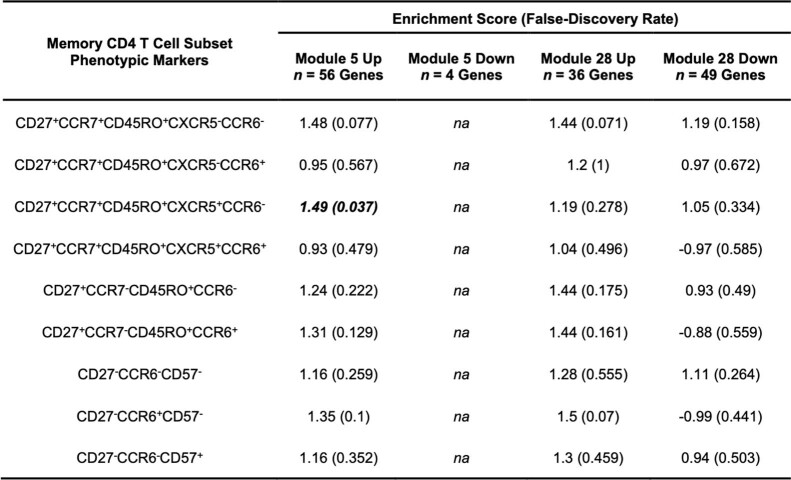
Extended Data Table 2.
Enrichment of Signature Genes within Sorted Memory CD4 T Cell Subsets from ART-Treated PWH
Naïve and memory CD4 T cell subsets were sorted from PBMC of 9 ART-treated PWH as shown in Extended Data Fig. 6 and were then subjected to standard RNA-seq. Transcriptomic signatures of HIV DNA+ memory CD4 T cells defined by WGCNA modules were compared with transcriptomes of memory CD4 T cell subsets using GSEA pre-ranked analysis as described in Methods. Each WGCNA module gene list was separated into two lists, one each for genes that were either higher (up) or lower (down) in HIV DNA+ memory CD4 T cells relative to HIV DNA− memory CD4 T cells. A false-discovery rate p ≤0.05 was considered to represent statistically significant enrichment. The enrichment score and false-discovery rate for the CD27+CCR7+CD45RO+CXCR5+CCR6− subset are shown in bold italics to indicate statistically significant enrichment for this subset. na, not attempted due to gene set size below minimum required for analysis.