Figure 3: Examples of recent modeling applications, from nucleosomes to chromosomes, and gene loci.
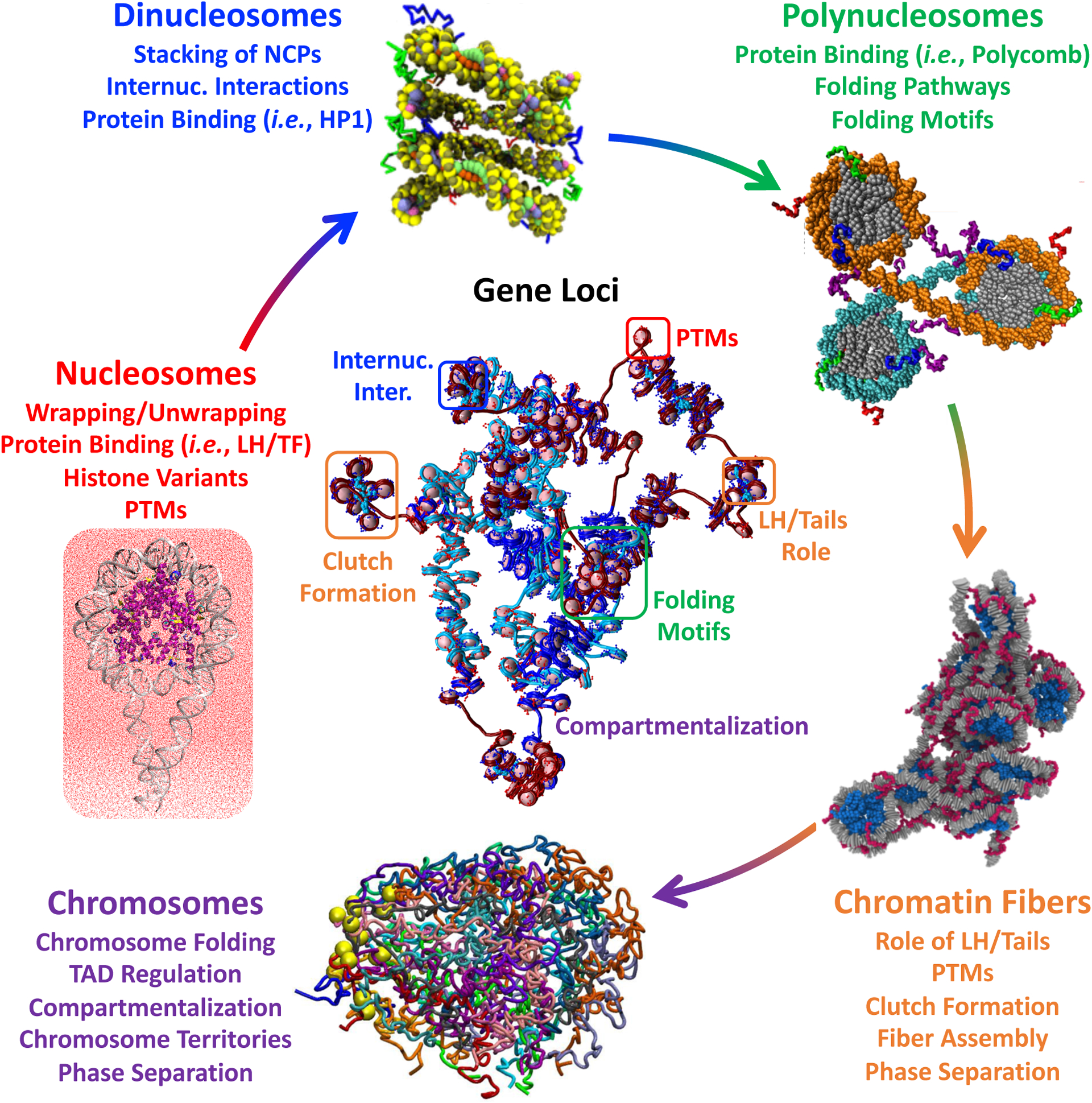
Nucleosome simulations provide insights into dynamical phenomena like wrapping and unwrapping, sliding, and breathing, the effect of LH and TF binding, and role of post translational modifications and histone variants. Dinucleosome systems are used to study regulation of internucleosme interactions and binding of repressing proteins like HP1. Polynucleosome studies reveal folding motifs and pathways, and the effect of repressive proteins like polycomb. Chromatin fibers at the kb-level reveal typologies, nucleosome clutches, role of linker DNA, LH, and histone tails, and phase separation. Modeling of chromosomes provide insights into folding and TAD formation/regulation, compartmentalization, chromosome territories, and phase separation. For gene loci, the elements and phenomena studied at each genomic scale are combined to obtain a high resolution structure. Molecular modeling images were adapted with permission from: chromatin fibers [50]** and chromosomes [142]** under Creative Commons Attribution License http://creativecommons.org/licenses/by/4.0/; dinucleosome [118] https://pubs.acs.org/doi/10.1021/acscentsci.1c00085 and under Creative Commons Attribution License https://creativecommons.org/licenses/by-nc-nd/4.0/, polynucleosome [119] Copyright 2022 Elsevier, 21 and gene loci [146]**.
